I use an implementation of a graph variational auto-encoder (GVAE) that is available at this link: “gae-pytorch/gae at master · zfjsail/gae-pytorch · GitHub”.
The adjacency matrix is derived from the graph data object. Since there are no node features, data.x is filled with ones (represents an identity matrix).
My code is:
DATA PRE-PROCESSING:
import networkx as nx
import torch_geometric
from torch_geometric.datasets import Planetoid
from scipy.sparse import coo_matrix, hstack, vstack
!pip install tensorboardX
from tensorboardX import SummaryWriter
data = GI_data_list[1]
print("graph data: ", data)
HPA_graph = nx.Graph()
edgeinfo = data.edge_index
src = edgeinfo[0].cpu().numpy()
dst = edgeinfo[1].cpu().numpy()
edgelist = zip(src,dst)
for i,j in edgelist:
HPA_graph.add_edge(i,j)
A = nx.adjacency_matrix(HPA_graph) # ADJACENCY MATRIX USED IN TRAINING
non_edges = list(nx.non_edges(HPA_graph))
e0 = [tup[0] for tup in non_edges]
e1 = [tup[1] for tup in non_edges]
neg_edge_list = np.vstack((e0, e1))
neg_edge_index = torch.as_tensor(neg_edge_list, dtype=torch.long)
GRAPH DATA FORMAT:
The adjacency matrix derived from the edge_index (edge list) has the shape (3163, 3163).
POSITIVE WEIGHT FACTOR:
w_factor = (neg_edge_index.shape[1]/edgeinfo.shape[1])
w_factor = 79.3
GRAPH CONVOLUTION CLASS:
import torch
import torch.nn.functional as F
from torch.nn.modules.module import Module
from torch.nn.parameter import Parameter
class GraphConvolution(Module):
def __init__(self, in_features, out_features, dropout):
super(GraphConvolution, self).__init__()
self.in_features = in_features
self.out_features = out_features
self.dropout = dropout
self.weight = Parameter(torch.DoubleTensor(in_features, out_features))
self.reset_parameters()
def reset_parameters(self):
torch.nn.init.xavier_uniform_(self.weight)
def forward(self, input, adj):
input = F.dropout(input, self.dropout, self.training)
support = torch.mm(input, self.weight)
output = torch.spmm(adj, support)
return output
def __repr__(self):
return self.__class__.__name__ + ' (' \
+ str(self.in_features) + ' -> ' \
+ str(self.out_features) + ')'
MODEL:
import torch
import torch.nn as nn
import torch.nn.functional as F
class GCNModelVAE(nn.Module):
def __init__(self, input_feat_dim, hidden_dim1, hidden_dim2, dropout):
super(GCNModelVAE, self).__init__()
self.gc1 = GraphConvolution(input_feat_dim, hidden_dim1, dropout)
self.gc2 = GraphConvolution(hidden_dim1, hidden_dim2, dropout)
self.gc3 = GraphConvolution(hidden_dim1, hidden_dim2, dropout)
self.dc = InnerProductDecoder(dropout)
def encode(self, x, adj):
hidden1 = F.relu(self.gc1(x, adj))
return self.gc2(hidden1, adj), self.gc3(hidden1, adj)
def reparameterize(self, mu, logvar):
if self.training:
std = torch.exp(logvar)
eps = torch.randn_like(std)
return eps.mul(std).add_(mu)
else:
return mu
def forward(self, x, adj):
mu, logvar = self.encode(x, adj)
z = self.reparameterize(mu, logvar)
return self.dc(z), mu, logvar
class InnerProductDecoder(nn.Module):
"""Decoder for using inner product for prediction."""
def __init__(self, dropout):
super(InnerProductDecoder, self).__init__()
self.dropout = dropout
self.act = torch.sigmoid
def forward(self, z):
z = F.dropout(z, self.dropout, training=self.training)
adj = self.act(torch.mm(z, z.t()))
return adj
LOSS FUNCTION:
import torch
import torch.nn.modules.loss
import torch.nn.functional as F
def loss_function(preds, labels, mu, logvar, n_nodes, norm, weight):
cost = norm * F.binary_cross_entropy_with_logits(preds, labels, pos_weight=weight)
KLD = -0.5 / n_nodes * torch.mean(torch.sum(
1 + 2 * logvar - mu.pow(2) - logvar.exp().pow(2), 1))
return cost + KLD
UTILS:
import pickle as pkl
import networkx as nx
import numpy as np
import scipy.sparse as sp
import torch
from sklearn.metrics import roc_auc_score, average_precision_score
from sklearn.metrics import confusion_matrix
from sklearn.metrics import f1_score
def sparse_to_tuple(sparse_mx):
if not sp.isspmatrix_coo(sparse_mx):
sparse_mx = sparse_mx.tocoo()
coords = np.vstack((sparse_mx.row, sparse_mx.col)).transpose()
values = sparse_mx.data
shape = sparse_mx.shape
return coords, values, shape
def mask_test_edges(adj):
# Function to build test set with 10% positive links
# Remove diagonal elements
adj = adj - sp.dia_matrix((adj.diagonal()[np.newaxis, :], [0]), shape=adj.shape)
adj.eliminate_zeros()
# Check that diag is zero:
# assert np.diag(adj.todense()).sum() == 0
adj_triu = sp.triu(adj)
adj_tuple = sparse_to_tuple(adj_triu)
edges = adj_tuple[0]
edges_all = sparse_to_tuple(adj)[0]
num_test = int(np.floor(edges.shape[0] / 10.))
num_val = int(np.floor(edges.shape[0] / 20.))
all_edge_idx = list(range(edges.shape[0]))
np.random.shuffle(all_edge_idx)
val_edge_idx = all_edge_idx[:num_val]
test_edge_idx = all_edge_idx[num_val:(num_val + num_test)]
test_edges = edges[test_edge_idx]
val_edges = edges[val_edge_idx]
train_edges = np.delete(edges, np.hstack([test_edge_idx, val_edge_idx]), axis=0)
def ismember(a, b, tol=5):
rows_close = np.all(np.round(a - b[:, None], tol) == 0, axis=-1)
return np.any(rows_close)
test_edges_false = []
while len(test_edges_false) < len(test_edges):
idx_i = np.random.randint(0, adj.shape[0])
idx_j = np.random.randint(0, adj.shape[0])
if idx_i == idx_j:
continue
if ismember([idx_i, idx_j], edges_all):
continue
if test_edges_false:
if ismember([idx_j, idx_i], np.array(test_edges_false)):
continue
if ismember([idx_i, idx_j], np.array(test_edges_false)):
continue
test_edges_false.append([idx_i, idx_j])
val_edges_false = []
while len(val_edges_false) < len(val_edges):
idx_i = np.random.randint(0, adj.shape[0])
idx_j = np.random.randint(0, adj.shape[0])
if idx_i == idx_j:
continue
if ismember([idx_i, idx_j], train_edges):
continue
if ismember([idx_j, idx_i], train_edges):
continue
if ismember([idx_i, idx_j], val_edges):
continue
if ismember([idx_j, idx_i], val_edges):
continue
if val_edges_false:
if ismember([idx_j, idx_i], np.array(val_edges_false)):
continue
if ismember([idx_i, idx_j], np.array(val_edges_false)):
continue
val_edges_false.append([idx_i, idx_j])
data = np.ones(train_edges.shape[0])
# Re-build adj matrix
adj_train = sp.csr_matrix((data, (train_edges[:, 0], train_edges[:, 1])), shape=adj.shape)
adj_train = adj_train + adj_train.T
# NOTE: these edge lists only contain single direction of edge!
return adj_train, train_edges, val_edges, val_edges_false, test_edges, test_edges_false
def preprocess_graph(adj):
adj = sp.coo_matrix(adj)
adj_ = adj + sp.eye(adj.shape[0])
rowsum = np.array(adj_.sum(1))
degree_mat_inv_sqrt = sp.diags(np.power(rowsum, -0.5).flatten())
adj_normalized = adj_.dot(degree_mat_inv_sqrt).transpose().dot(degree_mat_inv_sqrt).tocoo()
return sparse_mx_to_torch_sparse_tensor(adj_normalized)
def sparse_mx_to_torch_sparse_tensor(sparse_mx):
"""Convert a scipy sparse matrix to a torch sparse tensor."""
sparse_mx = sparse_mx.tocoo().astype(np.float64)
indices = torch.from_numpy(
np.vstack((sparse_mx.row, sparse_mx.col)).astype(np.int64))
values = torch.from_numpy(sparse_mx.data)
shape = torch.Size(sparse_mx.shape)
return torch.sparse.DoubleTensor(indices, values, shape)
def get_roc_score(emb, adj_orig, edges_pos, edges_neg):
def sigmoid(x):
x = np.float128(x)
return 1 / (1 + np.exp(-x))
# Predict on test set of edges
adj_rec = np.dot(emb, emb.T)
preds = []
pos = []
for e in edges_pos:
preds.append(sigmoid(adj_rec[e[0], e[1]]))
pos.append(adj_orig[e[0], e[1]])
preds_neg = []
neg = []
for e in edges_neg:
preds_neg.append(sigmoid(adj_rec[e[0], e[1]]))
neg.append(adj_orig[e[0], e[1]])
preds_all = np.hstack([preds, preds_neg])
labels_all = np.hstack([np.ones(len(preds)), np.zeros(len(preds))])
p = np.array([1 if p>0.5 else 0 for p in preds_all]).reshape((-1,1))
y = labels_all.reshape((-1,1))
roc_score = roc_auc_score(labels_all, preds_all)
ap_score = average_precision_score(labels_all, preds_all)
c = confusion_matrix(y, p, normalize='true')
# print("confusion matrix: ", c)
tn, fp, fn, tp = c.ravel()
# print("tn: ", tn, "fp: ", fp, "fn: ", fn, "tp: ", tp)
balanced_acc = (tp+tn)/2
f1_sco = f1_score(y, p, average='weighted')
return roc_score, ap_score, balanced_acc, f1_sco
TRAINING:
from __future__ import division
from __future__ import print_function
import argparse
import numpy as np
import scipy.sparse as sp
import torch
from torch import optim
seed = int(42)
hidden1 = int(256)
hidden2 = int(128)
lr = 0.01
dropout = np.float64(0)
adj = A
features = torch.as_tensor(data.x , dtype=torch.float64)
n_nodes, feat_dim = features.shape
#Store original adjacency matrix (without diagonal entries) for later
adj_orig = adj
adj_orig = adj_orig - sp.dia_matrix((adj_orig.diagonal()[np.newaxis, :], [0]), shape=adj_orig.shape)
adj_orig.eliminate_zeros()
adj_train, train_edges, val_edges, val_edges_false, test_edges, test_edges_false = mask_test_edges(adj)
adj = adj_train
#Some preprocessing
adj_norm = preprocess_graph(adj)
adj_label = adj_train + sp.eye(adj_train.shape[0])
adj_label = torch.DoubleTensor(adj_label.toarray())
norm = adj.shape[0] * adj.shape[0] / float((adj.shape[0] * adj.shape[0] - adj.sum()) * 2)
w = np.ones(adj_label.shape)
w = w*w_factor
pos_weight = torch.as_tensor(w, dtype=torch.float64)
hidden_emb = None
epochs = int(900)
model = GCNModelVAE(feat_dim, hidden1, hidden2, dropout)
optimizer = optim.Adam(model.parameters(), lr=lr)
for epoch in range(epochs):
t = time.time()
model.train()
optimizer.zero_grad()
recovered, mu, logvar = model(features, adj_norm)
loss = loss_function(preds=recovered, labels=adj_label,
mu=mu, logvar=logvar, n_nodes=n_nodes,
norm=norm, weight=pos_weight)
loss.backward()
cur_loss = loss.item()
optimizer.step()
hidden_emb = mu.data.numpy()
roc_curr, ap_curr, balanced_acc, f1_sco = get_roc_score(hidden_emb, adj_orig, val_edges, val_edges_false)
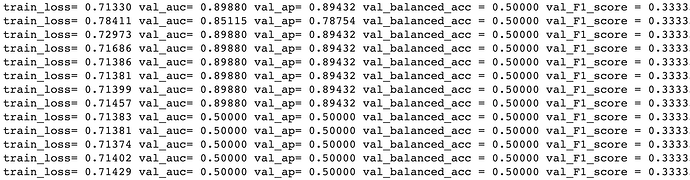
print("Epoch:", '%04d' % (epoch + 1), "train_loss=", "{:.5f}".format(cur_loss),
"val_auc=", "{:.5f}".format(roc_curr),
"val_ap=", "{:.5f}".format(ap_curr),
"val_balanced_acc =", "{:.5f}".format(balanced_acc),
"val_F1_score =", "{:.5f}".format(f1_sco),
"time=", "{:.5f}".format(time.time() - t)
)
print("Optimization Finished!")
roc_score, ap_score, balanced_acc, f1_sco = get_roc_score(hidden_emb, adj_orig, test_edges, test_edges_false)
print('Test ROC AUC score: ' + str(roc_score))
print('Test AP score: ' + str(ap_score))
print('Test Bal_acc score: ' + str(balanced_acc))
print('Test F1 score: ' + str(f1_sco))