You would have to move all tensors to the same device as was done for the model.
In particular mask has to be moved.
Thanks!
I solved it thanks to you 
mask.to(device)
Hello!
I would like to implement something like this dummy code (or as in the figure in the original post), but for an arbitrary number of sub-networks, maybe > 100, i.e.:
self.network1 = MySmallModel()
self.network2 = MySmallModel()
. .
. .
. .
self.network100 = MySmallModel()
Could you think of a pythonic way to implement this that might not involve writing 100+ lines of code?
Alternatively, is there something like the “groups” parameter of a Conv layer for a Linear layer?
Thanks a lot!
You could use an nn.ModuleList() or nn.ModuleDict and append each model to it as seen here:
modules = nn.ModuleList()
for _ in range(100):
modules.append(nn.Linear(1, 1))
modules = nn.ModuleDict()
for i in range(100):
modules['network{}'.format(i)] = nn.Linear(1, 1)
I don’t think so (at least I cannot think of a clean way to use a “grouped” linear approach). nn.Linear generally accepts inputs as [batch_size, *, nb_features], where the * defines arbitrary dimensions.
The linear layer will then be applied to each sample in these additional dimensions as if you would feed it in a loop to it.
Dear Mr. ptrblck,
First, I want to show my respect. You are really incredible.
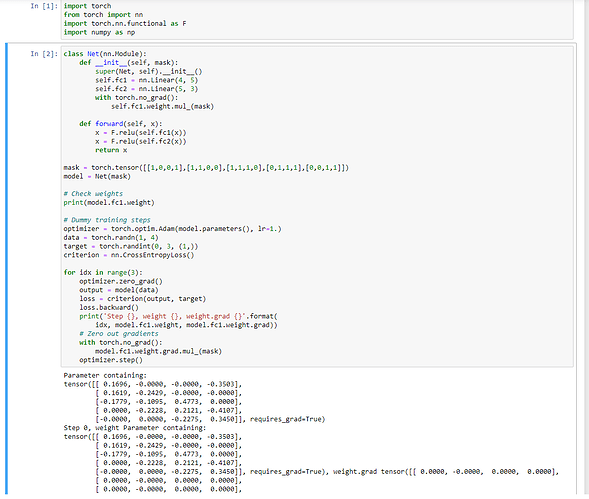
And I have run your code, but the result is far from my understanding, I thought in gradient, only the masked value will be zero, but there is only one non-zero row(some of the step is exactly all zero).
The result :
Step 0, weight Parameter containing:
tensor([[ 0.0998, 0.0000, -0.0000, -0.3424],
[ 0.0467, -0.4460, 0.0000, 0.0000],
[-0.3696, 0.1440, 0.1768, -0.0000],
[-0.0000, -0.3162, -0.3985, -0.4582],
[ 0.0000, 0.0000, 0.4535, -0.2398]], requires_grad=True),
weight.grad tensor([[ 0.0000, -0.0000, 0.0000, -0.0000],
[ 0.0000, -0.0000, 0.0000, -0.0000],
[ 0.0000, -0.0000, 0.0000, -0.0000],
[ 0.0000, -0.0000, 0.0000, -0.0000],
[ 0.1031, -0.0314, 0.0238, -0.0132]])
Then, I have another question.
If we just mask the gradient before optimizer(after “loss.backward()”), is the gradient back propagate through the non-connected edge? That is not I want.
I don’t know which code snippet to you running, so could you post the link or the code snippet you were using and explain your use case a bit more, please?
Autograd won’t backpropagate through unused parameters. However, masking the gradient does not disconnect the parameter from the computation graph. Since the gradients were already calculated (you would have to get the gradients before being able to mask them), I’m unsure if I understand your concern correctly.
Hello Mr.ptrblck,
First question:
This is the entire code I run, where the section 2 of my code is exactly yours from your answer on January 20.
The result is like the one I post before(zeros not appear in mask place only).
Second one:
I think that’s what I mean, but if I can’t disconnect the parameter by masking, what should I do to really disconnected it to prevent the influence of gradient.
Thank you for your reply.
In the provided code snippets, not only the gradients but also the fc1.weight is using the mask, which could create zero outputs and thus zero gradients on more indices than used in the mask.
The code snippet was created according to the original use case.
If you don’t want to train the parameter at all, you can set its requires_grad attribute to False.
Hi Mr. ptrblck,
I think I still not understand why the zero is in row-shape. If we have masked the weight first, we still get the gradient in the masked parameter right? So, they should still got gradient, after all, the input data is non-zero, which is the ratio that should be updated.
Second one:
I want the gradient influence through the connected link only(but not unconnected one), is that possible? How can I implement it, my best option is to use tons nn.Linear to create a layer. What I mean is like use the nn.Linear(n, 1) to create the node in the next layer (the connection of layer is fixed and is known). I don’t how to use the more direct way to do it like mask.
BTW, we should ignore the unconnected-link-gradient right? To update in the more accurate way?
Thank you for reply.
The zero gradients can not only be created by the masking, but also during the training itself depending on the model architecture.
You could remove e.g. the last relu and only mask a single value to verify this claim:
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.fc1 = nn.Linear(4, 5)
self.fc2 = nn.Linear(5, 3)
def forward(self, x):
x = F.sigmoid(self.fc1(x))
x = self.fc2(x)
return x
model = Net()
mask = torch.ones_like(model.fc1.weight)
mask[0, 0] = 0.
# Check weights
print(model.fc1.weight) # all valid
# Dummy training steps
optimizer = torch.optim.Adam(model.parameters(), lr=1.)
data = torch.randn(1, 4)
target = torch.randint(0, 3, (1,))
criterion = nn.CrossEntropyLoss()
for idx in range(3):
optimizer.zero_grad()
output = model(data)
loss = criterion(output, target)
loss.backward()
# Zero out gradients
with torch.no_grad():
model.fc1.weight.grad.mul_(mask)
print('Step {}, weight {}, weight.grad {}'.format(
idx, model.fc1.weight, model.fc1.weight.grad))
optimizer.step()
Here you can see that only a single gradient is masked and only this one is indeed set to zero.
If other gradients also drop to zero after some iterations, it’s not due to the masking.
I don’t know what “connected/unconnected link” means, so could you explain what you are trying to achieve?
Hello, Mr. ptrblck:
I understand nearly the whole story, thank you for your patient.
The reason I got so many zero at the first time is that you use relu function in first layer, which make the input of second layer is zero is that right?
I am now trying to learn using learning model to decoding.
And to decode, we don’t need fully connected, because there are some specific feature of the code.
Let me use some picture to explain:
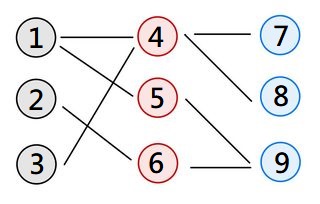
In this case, node 1 and node 4 is connected, and 4 and 9 is unconnected.
However, if I just mask the gradient like that the gradient from 4-9 edge will influence the update of 1-4 edge(I am not sure this is true or not). But that is not I want. Hope that is clear for you. I want to know how to implement like this.
Thank you for your reply.
Thanks for the update. I believe your use case comes close to the original question and you could mask the weight matrix as well as the gradients to achieve the custom pattern.
Alternatively, you could also take a look at sparse layers.
Mr. ptrblck
Thanks for your help, I really appreciate.