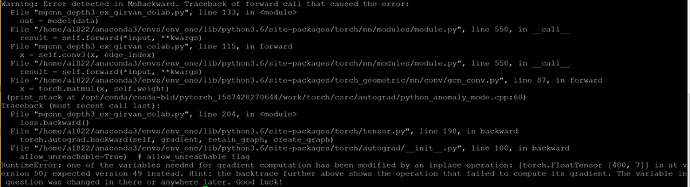
Can’t find the in-place operation anywhere? Tried replacing many variables with variable_temp… It comes from loss.backward here:
for epoch in range(10):
print(epoch)
for i, l in trainloader:
i_temp = i.to(device) # i added temps in this section 20:50 30.04.20
l_temp = l.to(device)
print(i_temp.size(), l_temp.size())
optimizer.zero_grad()
output = model(i_temp)
loss = criterion(output, l_temp)
print(output.size(), loss.size())
loss.backward(retain_graph=True)
optimizer.step()
Here is the full code for reference:
#!/usr/bin/env python3
import networkx as nx
import math
import csv
import random as rand
import sys
import matplotlib.pyplot as plt
from matplotlib import colors as mcolors
import torch_geometric
from torch_geometric.datasets import Planetoid
dataset = Planetoid(root='/local/scratch/al822/test', name='Cora')
G = torch_geometric.utils.to_networkx(dataset[0])
G = G.to_undirected()
#G.remove_nodes_from(range(2000,2709))
fig = plt.figure(1,figsize=(20,20))
nx.draw(G,node_size=60,font_size=8, with_labels=False, edge_color="white")
fig.set_facecolor("#00000F")
plt.show()
print(G.nodes())
G_list = nx.read_gpickle("g_list.gpickle")
print(G_list)
figtest = plt.figure(3,figsize=(30,30))
print(G_list[0].number_of_edges())
print(G_list[0].number_of_nodes())
nx.draw(G_list[0],node_size=10,font_size=8, with_labels=False)
plt.show()
pos=nx.spring_layout(G_list[0],k=3)
comms_1 = [{0}, {1, 2}, {3}, {4}, {5}, {6}, {208, 7}, {8, 281, 101, 269}, {9}, {10}, {11}, {12}, {13}, {158, 180, 14, 159}, {15}, {16}, {201, 297, 17, 24, 185}, {18, 139, 103}, {19}, {20}, {21}, {22, 39}, {23}, {25}, {99, 26, 123, 122}, {27}, {28}, {29}, {30}, {31}, {32, 242, 270, 279}, {33, 286}, {34}, {35}, {36}, {164, 37, 210, 211, 55, 60}, {38}, {40}, {41, 175}, {42, 87}, {152, 43}, {44}, {45}, {46}, {163, 47}, {48}, {49}, {50}, {51}, {52}, {53}, {54}, {56}, {57}, {58}, {105, 59}, {61}, {62}, {63}, {64}, {65, 239}, {66}, {282, 67}, {68}, {69}, {70}, {206, 71}, {72}, {73}, {74}, {75, 84, 284}, {88, 162, 76, 130}, {77}, {78}, {79}, {80, 257}, {81}, {82}, {83}, {85}, {86}, {89, 258}, {90, 155, 156}, {91}, {92}, {93}, {195, 94}, {95}, {96}, {97}, {98}, {100}, {289, 133, 102, 138, 236, 109, 153, 124, 126}, {104}, {106}, {107}, {108}, {110}, {111}, {112}, {113}, {114}, {115}, {116}, {259, 117}, {118, 255}, {119}, {120}, {121}, {125}, {127}, {128, 233}, {129}, {131}, {132}, {134}, {137, 135}, {136}, {140}, {141}, {142}, {143}, {144, 145, 213}, {146}, {147}, {148}, {149}, {150}, {151}, {154}, {157}, {160, 277}, {161}, {165}, {166, 271}, {168, 167}, {169}, {170}, {171}, {240, 172}, {173}, {174}, {197, 231, 232, 176, 179}, {177}, {178}, {181}, {182, 183}, {184}, {186}, {187}, {188}, {189}, {190}, {191}, {192}, {193}, {194}, {196}, {198}, {199}, {200}, {202}, {203}, {204}, {205}, {207}, {209}, {212}, {214}, {215}, {216}, {217, 243}, {218}, {219}, {220}, {221}, {222}, {223}, {224}, {225}, {226}, {227}, {228}, {229}, {230}, {234}, {235}, {237}, {238}, {241}, {244}, {245}, {246}, {247}, {248}, {249}, {250}, {251, 253}, {252}, {254}, {256}, {260}, {261}, {262}, {263}, {264}, {265}, {266}, {267}, {268}, {272}, {273}, {274}, {275}, {276}, {278}, {280}, {283}, {285}, {287}, {288}, {290}, {291}, {292}, {293}, {294}, {295}, {296}, {298}, {299}]
colors = dict(mcolors.BASE_COLORS, **mcolors.CSS4_COLORS)
fig2 = plt.figure(2,figsize=(20,20))
#for key, com in zip(colors.keys(), comms_1):
#com = list(map(str,com))
#nx.draw_networkx_nodes(G_list[0], pos,
#nodelist=com,
#node_color=key,
#node_size=50,
#alpha=1)
#nx.draw_networkx_edges(G_list[0],pos, width=0.5,alpha=1,edge_color='k')
# nx.draw_networkx_edges(G,pos,edgelist=removed_edges, width=2, edge_color='k')
#plt.show()
#nx.draw_networkx_edges(G,pos,edgelist=exist_edges, width=2,alpha=1,edge_color='k')
# nx.draw_networkx_edges(G,pos,edgelist=removed_edges, width=2, edge_color='k') #, style='dashed')
G_list[0].number_of_nodes()
G_list[0].nodes[4]
new_dataset = [None] * len(G_list)
#networkx removed all the features and other dataset keys, so have to reinsert them
for i in list(range(0,len(G_list))):
new_dataset[i] = torch_geometric.utils.from_networkx(G_list[i])
new_dataset[i].x = dataset[0].x
new_dataset[i].y = dataset[0].y
new_dataset[i].train_mask = dataset[0].train_mask
new_dataset[i].val_mask = dataset[0].val_mask
new_dataset[i].test_mask = dataset[0].test_mask
new_dataset[i].num_classes = dataset.num_classes
print(new_dataset)
new_dataset[0].num_node_features
dataset[0].train_mask[0:200]
new_dataset[0].test_mask
len(new_dataset)
new_dataset[0].train_mask.size()
import torch
import torch.nn.functional as F
from torch_geometric.nn import GCNConv
from torch import nn, optim
class GCNN(torch.nn.Module):
def __init__(self):
super(GCNN, self).__init__()
self.conv1 = GCNConv(new_dataset[0].num_node_features, 957)
self.conv2 = GCNConv(957, 480)
self.conv3 = GCNConv(480, new_dataset[0].num_classes)
def forward(self, data):
x, edge_index = data.x, data.edge_index
x = self.conv1(x, edge_index)
x = F.relu(x)
x = F.dropout(x, training=self.training)
x = self.conv2(x, edge_index)
x = F.relu(x)
x = F.dropout(x, training=self.training)
x = self.conv3(x, edge_index)
return F.log_softmax(x, dim=1)
out_list = []
for i in list(range(0,len(G_list)-1,200)): # -1 since last graph has no edges so GCNN cant work on it
print(i)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model = GCNN().to(device)
data = new_dataset[i].to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=0.01, weight_decay=5e-4)
model.train()
for epoch in range(50):
optimizer.zero_grad()
out = model(data)
print(out.size())
loss_gcn = F.nll_loss(out[data.train_mask], data.y[data.train_mask])
loss_gcn.backward(retain_graph=True) #change back to true
optimizer.step()
out_list.append(out)
print(out_list)
out_flat = torch.flatten(torch.cat(out_list, dim=0))
out_nodes = torch.split(out_flat, new_dataset[0].num_classes, dim=0)
labels = torch.cat([new_dataset[0].y]*(len(new_dataset)-1))
print(labels.size())
print(len(out_nodes))
labels_temp = labels.tolist()
train_data = []
for i in range(len(out_nodes)):
train_data.append([out_nodes[i], labels_temp[i]])
len(train_data)
from itertools import compress ############ filtering so only "train mask = true" nodes are put into training dataloader
train_data_temp = list(compress(train_data, (torch.cat([new_dataset[0].train_mask]*(len(new_dataset)-1))).tolist() )) # maybe in place
trainloader = torch.utils.data.DataLoader(train_data_temp, shuffle=True, batch_size=100)
i1, l1 = next(iter(trainloader))
print(l1.shape)
class FC(torch.nn.Module):
def __init__(self):
super(FC, self).__init__()
self.fc1 = nn.Linear(new_dataset[0].num_classes, new_dataset[0].num_classes)
self.fc2 = nn.Linear(new_dataset[0].num_classes, new_dataset[0].num_classes)
self.dropout = nn.Dropout(0.25)
self.dropout2 = nn.Dropout(0.25)
def forward(self, x):
# add dropout layer
x = self.dropout(x)
# add 1st hidden layer, with relu activation function
x = F.relu(self.fc1(x))
# add dropout layer
x = self.dropout2(x)
# add 2nd hidden layer, with relu activation function
x = self.fc2(x)
return F.log_softmax(x, dim=1)
#self.fc1 = nn.Linear(len(new_dataset) * new_dataset[0].num_nodes * new_dataset[0].num_classes, 500)
#self.fc2 = nn.Linear(500, new_dataset[0].num_nodes * new_dataset[0].num_classes)
model = FC().to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=0.01, weight_decay=5e-4)
criterion = nn.NLLLoss()
model.train()
for epoch in range(10):
print(epoch)
for i, l in trainloader:
i_temp = i.to(device) # i added temps in this section 20:50 30.04.20
l_temp = l.to(device)
print(i_temp.size(), l_temp.size())
optimizer.zero_grad()
output = model(i_temp)
loss = criterion(output, l_temp)
print(output.size(), loss.size())
loss.backward(retain_graph=True)
optimizer.step()
test_data = []
for i in range(len(out_nodes)):
test_data.append([out_nodes[i], labels_temp[i]])
from itertools import compress ############ filtering so only "test mask = true" nodes are put into testing dataloader
test_data = list(compress(test_data, (torch.cat([new_dataset[0].test_mask]*(len(new_dataset)-1))).tolist() ))
testloader = torch.utils.data.DataLoader(test_data, shuffle=True, batch_size=100)
model.eval()
correct = 0
for i, l in testloader:
i = i.to(device)
l = l.to(device)
pred = model(i).max(1)[1]
correct = correct + pred.eq(l).sum().item()
print(str(correct / len(test_data)))
file = open("mgcnn_depth2.txt","w")
file.write(str(correct / len(test_data)))
file.close()
RuntimeError Traceback (most recent call last)
in ()
15 loss = criterion(output, l_temp)
16 print(output.size(), loss.size())
—> 17 loss.backward(retain_graph=True)
18 optimizer.step()
19
1 frames
/usr/local/lib/python3.6/dist-packages/torch/autograd/init.py in backward(tensors, grad_tensors, retain_graph, create_graph, grad_variables)
98 Variable._execution_engine.run_backward(
99 tensors, grad_tensors, retain_graph, create_graph,
–> 100 allow_unreachable=True) # allow_unreachable flag
101
102
RuntimeError: one of the variables needed for gradient computation has been modified by an inplace operation: [torch.cuda.FloatTensor [480, 7]] is at version 50; expected version 49 instead. Hint: enable anomaly detection to find the operation that failed to compute its gradient, with torch.autograd.set_detect_anomaly(True).