I’ve been stuck here for a while now. Any help.
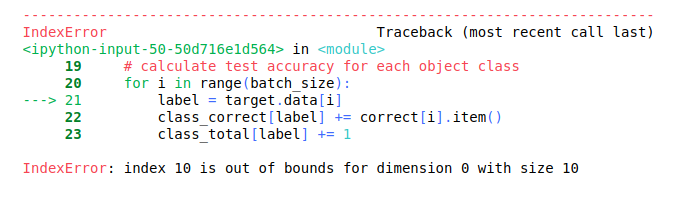
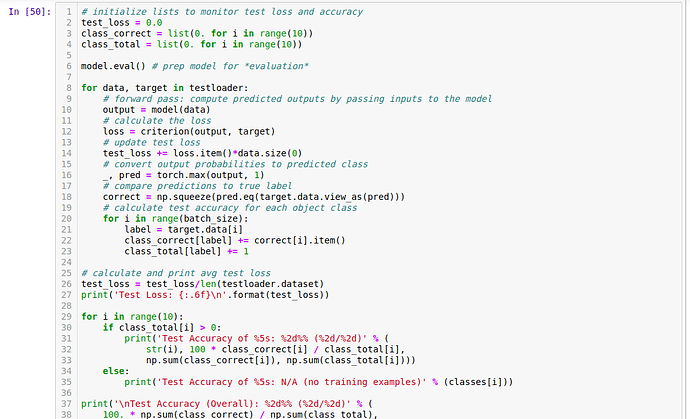
The last batch given by your DataLoader might be smaller, if the length of your Dataset is not divisible by the batch size without a remainder.
Try to get the current batch size inside the training loop:
for data, target in testloader:
batch_size = data.size(0)
...
for i in range(batch_size):
...
I worked perfectly!!!  Thank you!
Thank you!
Hi, I am having the same error. I understand that my code is trying to index outside of an allowed range, however I dont get how to fix this. I believe that edge_index is the cause. I attached my code and error, any help or direction would be appreciated.
class MoleculeDataset(Dataset):
def __init__(self, root,transform=None,test=False, pre_transform=None):
#root = data
""" Where the dataset should be stored. This folder is split
into raw_dir (downloaded dataset) and processed_dir (processed data).
"""
self.test = test
#self.filename = filename
super(MoleculeDataset, self).__init__(root, transform, pre_transform)
def process(self):
self.data = pd.read_csv(self.raw_paths[0])
for index, mol in tqdm(self.data.iterrows(), total=self.data.shape[0]):
mol_obj = Chem.MolFromSmiles(mol["SMILES"])
# Get node features
node_feats = self._get_node_features(mol_obj)
# Get edge features
# edge_feats = self._get_edge_features(mol_obj) un hash out to get edge feats
# Get adjacency info
edge_index = self._get_adjacency_info(mol_obj)
# Get labels info
label = self._get_labels(mol["Dev"])
# Create data object
data = Data(x=node_feats,
edge_index=edge_index,
#edge_attr=edge_feats, unhash me for edge feats
y=label,
smiles=mol["SMILES"]
)
if self.test:
torch.save(data,
os.path.join(self.processed_dir,
f'data_test_{index}.pt'))
else:
torch.save(data,
os.path.join(self.processed_dir,
f'data_{index}.pt'))
def _get_node_features(self, mol_array):
"""
This will return a matrix / 2d array of the shape
[Number of Nodes, Node Feature size]
"""
df = pd.read_csv('pka_model_302.csv')
smiles = df["SMILES"].values.tolist()
mol2 = [Chem.MolFromSmiles(sml) for sml in smiles]
mol_array = np.asarray(mol2)
for x in mol_array:
atom_features = get_atom_features(x)
func_features = functional(x)
combine_features = [atom_features.shape[0] // func_features.shape[0]] + [-1] * (len(func_features.shape) - 1)
atm_func_features = torch.cat((atom_features, func_features.expand(*combine_features)), dim=-1)
# atm_func_features_norm = min_max_scaler.fit_transform(atm_func_features)
#print(atm_func_features_norm.shape)
return torch.tensor(atm_func_features, dtype=torch.float) # 73 features, 10 atom specific. 63 "global"
def _get_adjacency_info(self, mol):
"""
We could also use rdmolops.GetAdjacencyMatrix(mol)
but we want to be sure that the order of the indices
matches the order of the edge features
"""
edge_indices = []
for bond in mol.GetBonds():
i = bond.GetBeginAtomIdx()
j = bond.GetEndAtomIdx()
edge_indices += [[i, j], [j, i]]
edge_indices = torch.tensor(edge_indices)
edge_indices = edge_indices.t().to(torch.long).view(2, -1)
return edge_indices
def _get_labels(self, label):
label = np.asarray([label])
return torch.tensor(label, dtype=torch.float)
def len(self):
return self.data.shape[0]
class GCN(torch.nn.Module):
def __init__(self):
# Init parent
super(GCN, self).__init__()
torch.manual_seed(42)
# 3 GCN layers. Learn info from 3 neighboor hops
self.initial_conv = GCNConv(atom_num_features, embedding_size)
self.conv1 = GCNConv(embedding_size, embedding_size)
self.conv1_bn = BatchNorm1d(embedding_size)
self.conv2 = GCNConv(embedding_size, embedding_size)
self.conv2_bn = BatchNorm1d(embedding_size)
self.conv3 = GCNConv(embedding_size, embedding_size)
self.conv3_bn = BatchNorm1d(embedding_size)
# self.dropout = Dropout(0.20)
# Output layer
self.out = Linear(embedding_size*2,1)
def forward(self, x, edge_index, batch_index):
# First Conv layer
hidden = self.initial_conv(x, edge_index)
hidden = tanh(hidden)
# Other Conv layers
hidden = self.conv1(hidden, edge_index)
hidden = tanh(self.conv1_bn(hidden))
hidden = self.conv2(hidden, edge_index)
hidden = tanh(self.conv2_bn(hidden))
hidden = self.conv3(hidden, edge_index)
hidden = tanh(self.conv3_bn(hidden))
# hidden = self.dropout(hidden)
# Global Pooling (stack different aggregations)
hidden = torch.cat([gmp(hidden, batch_index),
gap(hidden, batch_index)], dim=1)
# Apply a final (linear) classifier.
out = self.out(hidden)
return out,hidden
``
Traceback (most recent call last):
File “/home/alecsanc/Documents/descriptors/novartis/graphNN/complex_pytorch/dataset.py”, line 277, in
pred, embedding = model(batch.x.float(), batch.edge_index, batch.batch)
File “/home/alecsanc/anaconda3/envs/ml/lib/python3.9/site-packages/torch/nn/modules/module.py”, line 1110, in _call_impl
return forward_call(*input, **kwargs)
File “/home/alecsanc/Documents/descriptors/novartis/graphNN/complex_pytorch/dataset.py”, line 218, in forward
hidden = self.initial_conv(x, edge_index)
File “/home/alecsanc/anaconda3/envs/ml/lib/python3.9/site-packages/torch/nn/modules/module.py”, line 1110, in call_impl
return forward_call(*input, **kwargs)
File “/home/alecsanc/anaconda3/envs/ml/lib/python3.9/site-packages/torch_geometric/nn/conv/gcn_conv.py”, line 172, in forward
edge_index, edge_weight = gcn_norm( # yapf: disable
File “/home/alecsanc/anaconda3/envs/ml/lib/python3.9/site-packages/torch_geometric/nn/conv/gcn_conv.py”, line 64, in gcn_norm
deg = scatter_add(edge_weight, col, dim=0, dim_size=num_nodes)
File “/home/alecsanc/anaconda3/envs/ml/lib/python3.9/site-packages/torch_scatter/scatter.py”, line 29, in scatter_add
return scatter_sum(src, index, dim, out, dim_size)
File “/home/alecsanc/anaconda3/envs/ml/lib/python3.9/site-packages/torch_scatter/scatter.py”, line 21, in scatter_sum
return out.scatter_add(dim, index, src)
RuntimeError: index 26 is out of bounds for dimension 0 with size 26
Yes, I think your description is right and the indexing operation fails here:
return out.scatter_add(dim, index, src)
RuntimeError: index 26 is out of bounds for dimension 0 with size 26
This operation seems to be called by torch_scatter.scatter_add and indicates that an index with the value of 26 is used to index another tensor with a size of 26. This will fail, as an index of 26 expects a size of >=27 so maybe your indices are off by one?