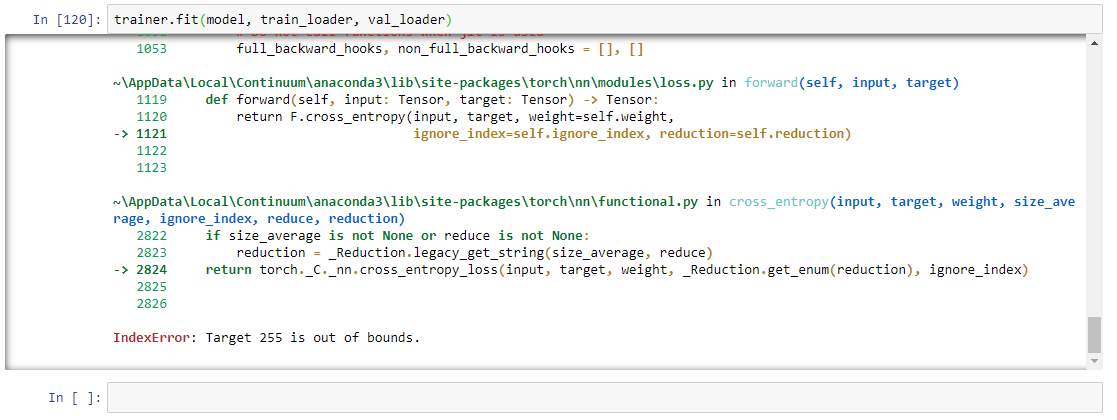
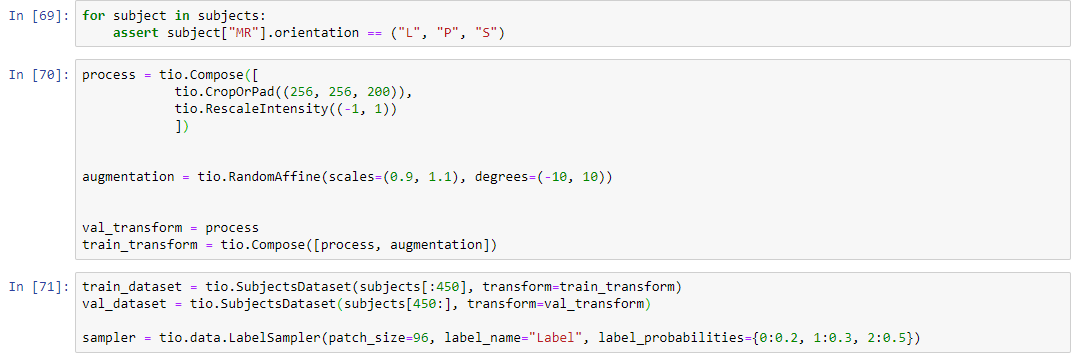
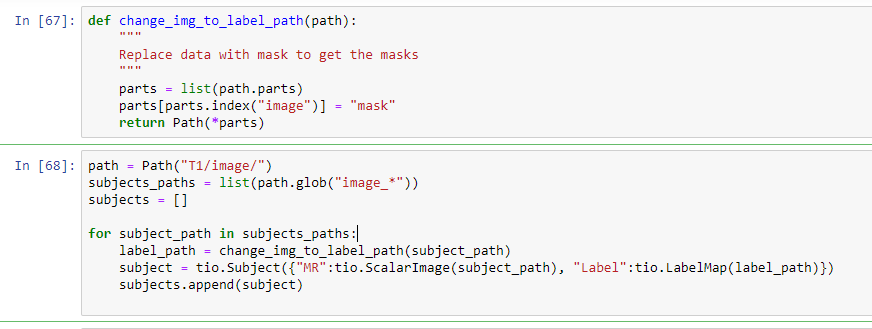
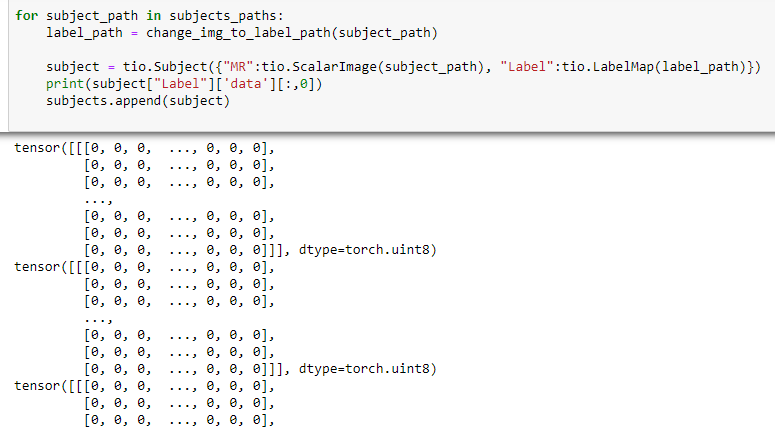
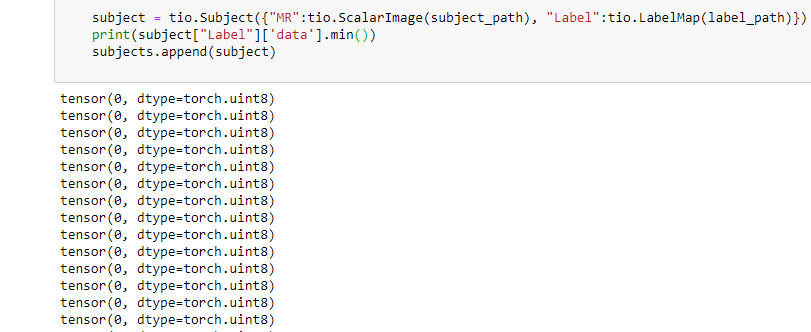
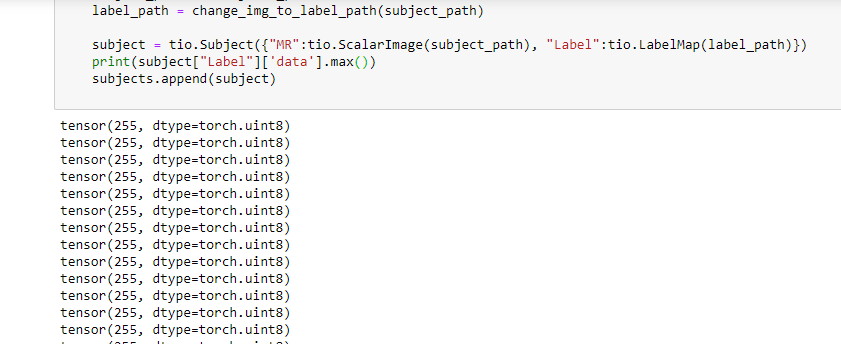
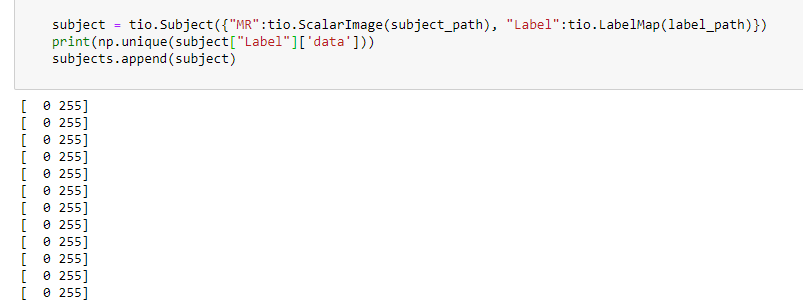
i upload the full code. maybe then you can understand.
def change_img_to_label_path(path):
"""
Replace data with mask to get the masks
"""
parts = list(path.parts)
parts[parts.index("image")] = "mask"
return Path(*parts)
path = Path("T1/image/")
subjects_paths = list(path.glob("image_*"))
subjects = []
for subject_path in subjects_paths:
label_path = change_img_to_label_path(subject_path)
subject = tio.Subject({"MR":tio.ScalarImage(subject_path), "Label":tio.LabelMap(label_path)})
subjects.append(subject)
for subject in subjects:
assert subject["MR"].orientation == ("L", "P", "S")
process = tio.Compose([
tio.CropOrPad((256, 256, 200)),
tio.RescaleIntensity((-1, 1))
])
augmentation = tio.RandomAffine(scales=(0.9, 1.1), degrees=(-10, 10))
val_transform = process
train_transform = tio.Compose([process, augmentation])
train_dataset = tio.SubjectsDataset(subjects[:650], transform=train_transform)
val_dataset = tio.SubjectsDataset(subjects[650:], transform=val_transform)
sampler = tio.data.LabelSampler(patch_size=96, label_name="Label", label_probabilities={0:0.2, 1:0.3, 2:0.5})
train_patches_queue = tio.Queue(
train_dataset,
max_length=40,
samples_per_volume=5,
sampler=sampler,
num_workers=4,
)
val_patches_queue = tio.Queue(
val_dataset,
max_length=40,
samples_per_volume=5,
sampler=sampler,
num_workers=4,
)
batch_size = 2
train_loader = torch.utils.data.DataLoader(train_patches_queue, batch_size=batch_size, num_workers=0)
val_loader = torch.utils.data.DataLoader(val_patches_queue, batch_size=batch_size, num_workers=0)
class Segmenter(pl.LightningModule):
def __init__(self):
super().__init__()
self.model = UNet()
self.optimizer = torch.optim.Adam(self.model.parameters(), lr=1e-4)
self.loss_fn = torch.nn.CrossEntropyLoss()
def forward(self, data):
pred = self.model(data)
return pred
def training_step(self, batch, batch_idx):
# You can obtain the raw volume arrays by accessing the data attribute of the subject
img = batch["MR"]["data"]
mask = batch["Label"]["data"][:,0] # Remove single channel as CrossEntropyLoss expects NxHxW
mask = mask.long()
pred = self(img)
loss = self.loss_fn(pred, mask)
# Logs
self.log("Train Loss", loss)
if batch_idx % 50 == 0:
self.log_images(img.cpu(), pred.cpu(), mask.cpu(), "Train")
return loss
def validation_step(self, batch, batch_idx):
# You can obtain the raw volume arrays by accessing the data attribute of the subject
img = batch["MR"]["data"]
mask = batch["Label"]["data"][:,0] # Remove single channel as CrossEntropyLoss expects NxHxW
mask = mask.long()
pred = self(img)
loss = self.loss_fn(pred, mask)
# Logs
self.log("Val Loss", loss)
self.log_images(img.cpu(), pred.cpu(), mask.cpu(), "Val")
return loss
def log_images(self, img, pred, mask, name):
results = []
pred = torch.argmax(pred, 1) # Take the output with the highest value
axial_slice = 50 # Always plot slice 50 of the 96 slices
fig, axis = plt.subplots(1, 2)
axis[0].imshow(img[0][0][:,:,axial_slice], cmap="bone")
mask_ = np.ma.masked_where(mask[0][:,:,axial_slice]==0, mask[0][:,:,axial_slice])
axis[0].imshow(mask_, alpha=0.6)
axis[0].set_title("Ground Truth")
axis[1].imshow(img[0][0][:,:,axial_slice], cmap="bone")
mask_ = np.ma.masked_where(pred[0][:,:,axial_slice]==0, pred[0][:,:,axial_slice])
axis[1].imshow(mask_, alpha=0.6, cmap="autumn")
axis[1].set_title("Pred")
self.logger.experiment.add_figure(f"{name} Prediction vs Label", fig, self.global_step)
def configure_optimizers(self):
#Caution! You always need to return a list here (just pack your optimizer into one :))
return [self.optimizer]