import torch
class TwoLayerNet(torch.nn.Module):
def __init__(self, D_in, H, D_out):
"""
In the constructor we instantiate two nn.Linear modules and assign them as
member variables.
"""
super(TwoLayerNet, self).__init__()
self.linear1 = torch.nn.Linear(D_in, H)
self.linear2 = torch.nn.Linear(H, D_out)
def forward(self, x):
"""
In the forward function we accept a Tensor of input data and we must return
a Tensor of output data. We can use Modules defined in the constructor as
well as arbitrary operators on Tensors.
"""
h_relu = self.linear1(x).clamp(min=0)
y_pred = self.linear2(h_relu)
return y_pred
dtype = torch.float
device = torch.device("cuda:0")
# N is batch size; D_in is input dimension;
# H is hidden dimension; D_out is output dimension.
N, D_in, H, D_out = 2140, 9216, 100, 30
def train(model,x,y,criterion,optimizer):
"""
train NN
"""
model.train()
y_pred = model(x)
loss = criterion(y_pred, y)
print('train-loss',t, loss.item(),end=' ')
optimizer.zero_grad()
loss.backward()
optimizer.step()
return loss.item()
def valid(model,x_valid,y_valid,criterion):
"""
test NN
"""
model.eval()
y_pred = model(x_valid)
loss = criterion(y_pred, y_valid)
print('test-loss',t, loss.item(),end=' ')
return loss.item()
# Create random Tensors to hold inputs and outputs
x_train = torch.tensor(torch.from_numpy(X_train),device=device,dtype=dtype)
y_train = torch.tensor(torch.from_numpy(y_train),device=device,dtype=dtype)
x_valid = torch.tensor(torch.from_numpy(X_valid),device=device,dtype=dtype)
y_valid = torch.tensor(torch.from_numpy(y_valid),device=device,dtype=dtype)
model = TwoLayerNet(D_in, H, D_out).to(device)
loss_train=[]
loss_valid=[]
criterion = torch.nn.MSELoss()
optimizer = torch.optim.SGD(model.parameters(), lr=1e-4)
for t in range(800):
loss_train.append(train(model,x_train,y_train,criterion,optimizer))
loss_valid.append(valid(model,x_valid,y_valid,criterion))
print()
Your code look alright.
What do you mean by “the same”?
Are both going down in the same manner, i.e. staying close to each other, or are both giving a constant value?
PS: I’ve edited your code for better readability. You can add code using three backticks` 
thanks for making code readable,they are going down in same manner i am not able to understand why, i have attached sample image
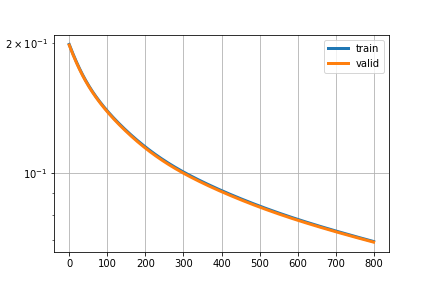
That’s pretty much exactly what you would want.
If the gap between the train and validation loss is really close you have low variance, which is great! Also it looks like the loss is going down, so you might want to continue the training.
Why do you expect something other to see? Should it be harder for the model to learn, i.e. did you expect a higher loss?
I’m not sure how you’ve loaded the data, but make sure the training and validation data is not the same.
ya variance should be close but validation loss should be greater and it is less than train loss, following is code for loading and splitting dataset
import os
import numpy as np
from pandas.io.parsers import read_csv
from sklearn.utils import shuffle
FTRAIN = 'all/training/training.csv'
FTEST = 'all/test/test.csv'
def load(test=False, cols=None):
"""Loads data from FTEST if *test* is True, otherwise from FTRAIN.
Pass a list of *cols* if you're only interested in a subset of the
target columns.
"""
fname = FTEST if test else FTRAIN
df = read_csv(os.path.expanduser(fname)) # load pandas dataframe
# The Image column has pixel values separated by space; convert
# the values to numpy arrays:
df['Image'] = df['Image'].apply(lambda im: np.fromstring(im, sep=' '))
if cols: # get a subset of columns
df = df[list(cols) + ['Image']]
print(df.count()) # prints the number of values for each column
df = df.dropna() # drop all rows that have missing values in them
X = np.vstack(df['Image'].values) / 255. # scale pixel values to [0, 1]
X = X.astype(np.float32)
if not test: # only FTRAIN has any target columns
y = df[df.columns[:-1]].values
y = (y - 48) / 48 # scale target coordinates to [-1, 1]
X, y = shuffle(X, y, random_state=42) # shuffle train data
y = y.astype(np.float32)
else:
y = None
return X, y
X, y = load()
print("X.shape == {}; X.min == {:.3f}; X.max == {:.3f}".format(
X.shape, X.min(), X.max()))
print("y.shape == {}; y.min == {:.3f}; y.max == {:.3f}".format(
y.shape, y.min(), y.max()))
import numpy as np
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split( X, y, test_size=0.2, random_state=42)
Blockquote
It’s a wild guess, but based on your loading function it looks like you are dealing with Kaggle’s facial keypoint detection dataset.
I’m not sure if there are multiple images of the same individuals in the training data, but I’m quite sure there are. Let’s assume your splitting gives two datasets which have nearly the same distribution (same individuals, gender ratio etc.).
Your training loss is currently saved before each optimizer step, while your validation loss is saved after the optimization is done. If both datasets are nearly identical, your validation loss might thus be a little lower if you don’t overfit your data.
Besides that I can’t find any other mistakes.
Are you following some kind of tutorial which assumes another validation loss curve?
ur wild guess is right, i was following Daniel’s tutorial http://danielnouri.org/notes/2014/12/17/using-convolutional-neural-nets-to-detect-facial-keypoints-tutorial/ where he has implemented it using Lasagne and Theano, I was trying to implement it using pytorch , he has also implemented two layer NN and his validation curve and training are quite different from mine.
Thanks for giving me your time
Ah OK. Let me check my code with yours. I remember reimplementing it some time ago in PyTorch using his blog post.
I have 4gb ram ,2gb ram gpu and when i am trying lenet-5 for same dataset i m getting
RuntimeError: CUDA error: out of memory. What should I do and what is causing this?
import torch
import torch.nn as nn
import torch.nn.functional as F
class LeNet(nn.Module):
def __init__(self):
super(LeNet, self).__init__()
# 1 input image channel, 6 output channels, 5x5 square convolution
# kernel
self.conv1 = nn.Conv2d(1, 32, 3)
self.conv2 = nn.Conv2d(6, 64, 2)
self.conv3 = nn.Conv2d(64, 128, 2)
self.fc1 = nn.Linear(128 * 5 * 5, 500)
self.fc2 = nn.Linear(500, 500)
self.fc3 = nn.Linear(500, 30)
def forward(self, x):
# Max pooling over a (2, 2) window
x = F.max_pool2d(F.relu(self.conv1(x)), (2, 2))
# If the size is a square you can only specify a single number
x = F.max_pool2d(F.relu(self.conv2(x)), (2, 2))
x = F.max_pool2d(F.relu(self.conv3(x)), (2, 2))
x = x.view(-1, self.num_flat_features(x))
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = F.relu(self.fc3(x))
x = self.fc3(x)
return x
def num_flat_features(self, x):
size = x.size()[1:] # all dimensions except the batch dimension
num_features = 1
for s in size:
num_features *= s
return num_features
dtype = torch.float
device = torch.device("cuda:0")
# N is batch size; D_in is input dimension;
# H is hidden dimension; D_out is output dimension.
N, D_in, H, D_out = 2140, 9216, 100, 30
def train(model,x,y,criterion,optimizer):
model.train()
y_pred = model(x)
loss = criterion(y_pred, y)
print('train-loss',t, loss.item(),end=' ')
optimizer.zero_grad()
loss.backward()
optimizer.step()
return loss.item()
def valid(model,x_valid,y_valid,criterion):
model.eval()
y_pred = model(x_valid)
loss = criterion(y_pred, y_valid)
print('test-loss',t, loss.item(),end=' ')
return loss.item()
# Create random Tensors to hold inputs and outputs
X_train=X_train.reshape(-1, 1, 96, 96)
X_valid=X_valid.reshape(-1, 1, 96, 96)
x_train = torch.tensor(torch.from_numpy(X_train),device=device,dtype=dtype)
y_train = torch.tensor(torch.from_numpy(Y_train),device=device,dtype=dtype)
x_valid = torch.tensor(torch.from_numpy(X_valid),device=device,dtype=dtype)
y_valid = torch.tensor(torch.from_numpy(Y_valid),device=device,dtype=dtype)
model = LeNet().to(device)
loss_train=[]
loss_valid=[]
criterion = torch.nn.MSELoss()
optimizer = torch.optim.SGD(model.parameters(), lr=1e-4)
for t in range(400):
loss_train.append(train(model,x_train,y_train,criterion,optimizer))
loss_valid.append(valid(model,x_valid,y_valid,criterion))
print()
Using your data split and my training code I got these loss curves:
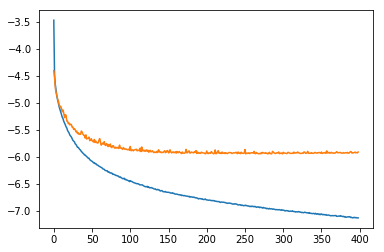
I used the same optimizer hyperparameters as in the blog, i.e. optim.SGD(lr=1e-2, momentum=0.9, nesterov=True). Also I initialized the parameters using xavier_uniform.
Your learning rate might just be too low for the FullyConnected model.
Hi ptrblck! I am facing the same trouble as mentioned in the post. My test loss is lower than the training loss. Can you please share the training code you used to reproduce the graph you posted here?
There might be some reasons for the test loss being lower than the training loss.
E.g. if you regularize your model with Dropout this is a common observation.
Is this the case in your use case?
This code should recreate approx. the same figure.