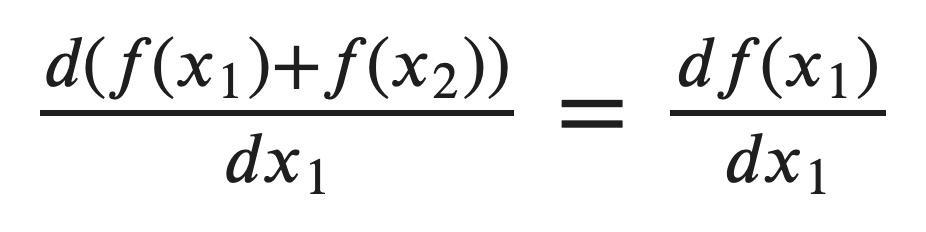I am hoping to get Jacobians in a way that respects the batch, efficiently
Given a batch of b (vector) predictions y_1,…,y_b, and inputs x_1 … x_b, I want to compute the Jacobians of y_i wrt x_i. In other words, I want a Jacobian of the output wrt input for each pair in the batch.
One might try the following:
import torch
import torch.nn as nn
# Load the experimental api
# https://github.com/pytorch/pytorch/blob/master/torch/autograd/functional.py
from experimental_api import jacobian
in_dim = 5
batch_size = 3
hidden_dim = 2
out_dim = 10
f = nn.Sequential(nn.Linear(in_dim, hidden_dim), nn.Sigmoid(), nn.Linear(hidden_dim, out_dim))
x = torch.randn(batch_size, in_dim)
result = jacobian(f, x)
result.shape
torch.Size([3, 10, 3, 5])
In this case, I wanted a result with shape (batch_size, out_dim, in_dim) = (3, 10, 5) but instead got extra dimensions. In fact, PyTorch is considering x as just one input matrix rather than a batch of several vectors. This leads to redundant calculation such as the derivatives of target 2 wrt input 1.
Clearly, I could do a loop like jacobian(f, x_row) for x_row in x but that would no longer use the GPU effectively. Can anyone propose an efficient solution?