Hi,
I am trying to write an LSTM Auto-encoder architecture to determine the reconstruction error for given multi-variate sequence. The purpose is to identify observations(rows) which are poorly reconstructed.
I am trying to wrap my head around the network mentioned below but couldn’t decide on how to write decoder network which takes input from encoder output and tries to reconstruct the input
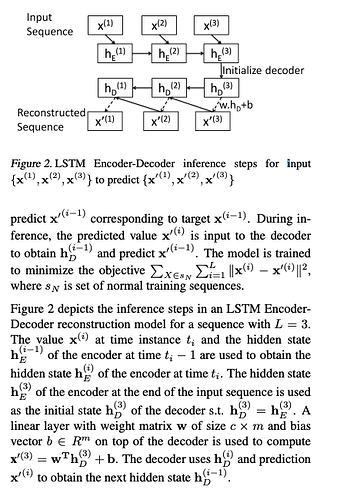
The architecture and its description is available at https://arxiv.org/pdf/1607.00148.pdf
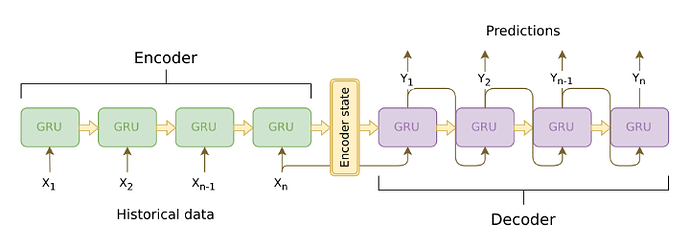
Similar architecture but with better visuals is
which is available at Joseph Eddy – Data science team leader and mentor, machine learning specialistclass LstmAutoEncoder(nn.module):
def __init__(self, x_dim, h_dim=(32, 16), z_dim=8, seq_length=144,
num_layers=1, dropout_frac=0.25, batchnorm=False):
super(LstmAutoEncoder, self).__init__()
self.x_dim = x_dim
self.h_dim = list(h_dim)
self.z_dim = z_dim
self.sq_l = seq_length
self.num_layers = num_layers
self.dropout_frac = dropout_frac
self.batchnorm = batchnorm
self.encoder = EncoderRNN(x_dim, h_dim, z_dim, num_layers, dropout_frac, batchnorm)
self.decoder = DecoderRNN(x_dim, h_dim, z_dim, num_layers, dropout_frac, batchnorm)
def forward(self, x):
"""
"""
z = self.encoder(x)
recon_x = self.decoder(z)
return recon_x
class EncoderRNN(nn.Module):
def __init__(self, x_dim, h_dim, z_dim, num_layers,
dropout_frac, batchnorm):
super(EncoderRNN, self).__init__()
self.nl = num_layers
self.drpt_fr = dropout_frac
self.bn = batchnorm
neurons = [x_dim, *h_dim, z_dim]
layers = [nn.LSTM(neurons[i - 1], neurons[i], self.nl, batch_first=True)
for i in range(1, len(neurons))]
self.hidden = nn.ModuleList(layers)
if self.bn:
bn_layers = [nn.BatchNorm1d(neurons[i]) for i in range(1, len(neurons))]
self.bns = nn.ModuleList(bn_layers)
def forward(self, x):
if self.bn:
for layer, bnm in zip(self.hidden, self.bns):
out, (hs, cs) = layer(x)
x = bnm(out)
x = nn.Dropout(p=self.drpt_fr)(x)
else:
for layer in self.hidden:
out, (hs, cs) = layer(x)
x = nn.Dropout(p=self.drpt_fr)(out)
return x[-1] # -1 is used to get only the last state as per the architecture in the pic
class DecoderRNN(nn.Module):
def __init__(self, x_dim, h_dim, z_dim, num_layers, dropout_frac, batchnorm):
super(DecoderRNN, self).__init__()
self.nl = num_layers
self.drpt_fr = dropout_frac
self.bn = batchnorm
h_dim = list(reversed(h_dim))
neurons = [z_dim] + h_dim
layers = [nn.LSTM(neurons[i - 1], neurons[i], self.nl, batch_first=True)
for i in range(1, len(neurons))]
self.hidden = nn.ModuleList(layers)
if batchnorm:
bn_layers = [nn.BatchNorm1d(neurons[i]) for i in range(1, len(neurons))]
self.bns = nn.ModuleList(bn_layers)
self.reconstruction = nn.Linear(h_dim[-1], x_dim)
def forward(self, x):
## this is the part I have trouble trying to code