I have been troubled by the missing of parameters after I did concatenation. I have read a similar post Model.named_parameters() will lose some layer modules - #5 by kaiyuyue, but the cause of my problem is different.
The Pytorch version is 0.3.1.post2.
The concatenation is as follows:
x = Variable(torch.cat((x.data, max_val), 1))
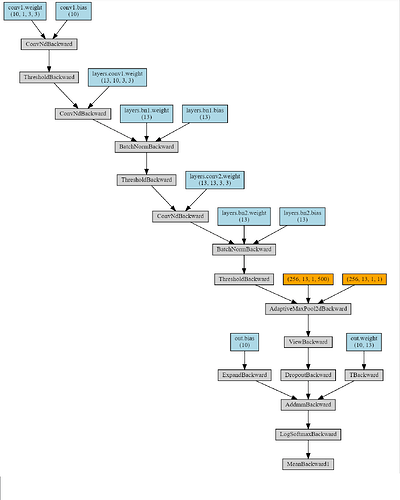
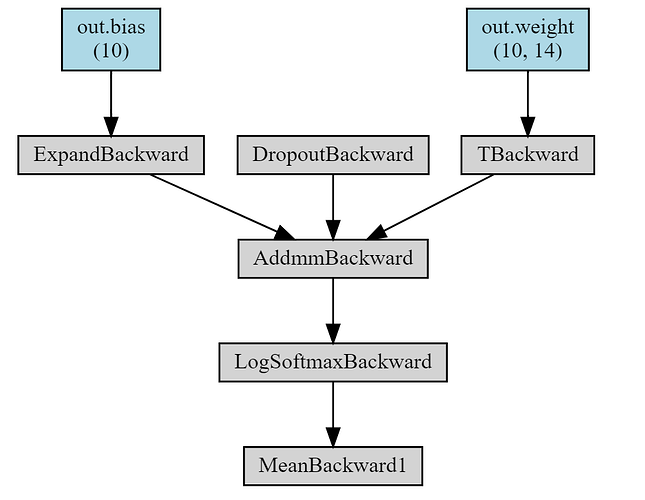
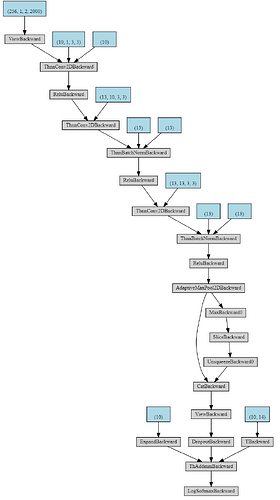
If I comment it out, the make_dot from torchviz shows all the parameters of the neural network. But if I execute the above line, most of the parameters are cut off and only the parameters of the last layer are shown in the diagram. Even if I make max_val a zero tensor (of the same size), the same cut off happens. The cut off caused a severe degradation in performance (prediction accuracy, the code not shown here) when applying the optimizer. Any hint? The code is below. Thanks.
%reload_ext autoreload
%autoreload 2
import os, re, math, copy, datetime, sys, traceback
import numpy as np
import scipy.signal as signal
import torch
from torch.autograd import Variable
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import Dataset, DataLoader
from torch.optim import lr_scheduler
import GPUtil
import pdb
gpu_avail = GPUtil.getAvailable()
if gpu_avail == []:
print ('There are no free GPUs; using CPU.')
use_gpu = False
else:
use_gpu = True
num_samples = 2000
numCats = 10
batch_size = 256
class BnLayer2(nn.Module):
def __init__(self, ni, nf, stride=2, kernel_size=(3,3)):
super().__init__()
self.conv1 = nn.Conv2d(ni, nf, kernel_size=kernel_size, stride=stride,
bias=False, padding=(1,1))
self.bn1 = nn.BatchNorm2d(nf)
self.conv2 = nn.Conv2d(nf, nf, kernel_size=kernel_size, stride=stride,
bias=False, padding=(1,1))
self.bn2 = nn.BatchNorm2d(nf)
def forward(self, x):
x = F.relu(self.bn1(self.conv1(x)))
x = F.relu(self.bn2(self.conv2(x)))
return x
class SimpleNet(nn.Module):
def __init__(self, layers, c, input_shape, p=0.2):
super().__init__()
self.input_shape = input_shape
self.conv1 = nn.Conv2d(1, layers[0], kernel_size=(3,3), stride=1, padding=(1,1))
self.layers = BnLayer2(layers[0], layers[1])
#self.out = nn.Linear(layers[-1] + 1, c) # with concatenation
self.out = nn.Linear(layers[-1], c) # without concatenation
self.drop = nn.Dropout(p)
def do_sth(self, x):
max_x = torch.max(x, 1)
return max_x
def forward(self, x):
x_init = x
x = x.view(-1,*self.input_shape)
x = F.relu(self.conv1(x))
x = self.layers(x)
x = F.adaptive_max_pool2d(x, 1)
max_x = self.do_sth(x)
max_val = torch.FloatTensor(max_x[0].data)
max_val = max_val[:, None, :, :] * 0
x = Variable(torch.cat((x.data, max_val), 1)) # concatenation causes parameters cut off
x = x.view(x.size(0), -1)
x = self.drop(x)
return F.log_softmax(self.out(x), dim=-1)
model = SimpleNet([10,13], numCats, (1,2,num_samples), p=0.20) # p = dropout
from graphviz import Digraph
from torchviz import make_dot
x=Variable(torch.randn(batch_size,1,2,num_samples))
y=model(x)
dot = make_dot(y.mean(),params=dict(model.named_parameters()))
dot.attr(size="140,140")
dot
The neural network without concatenation:
The neural network with concatenation (only the final layer is kept):