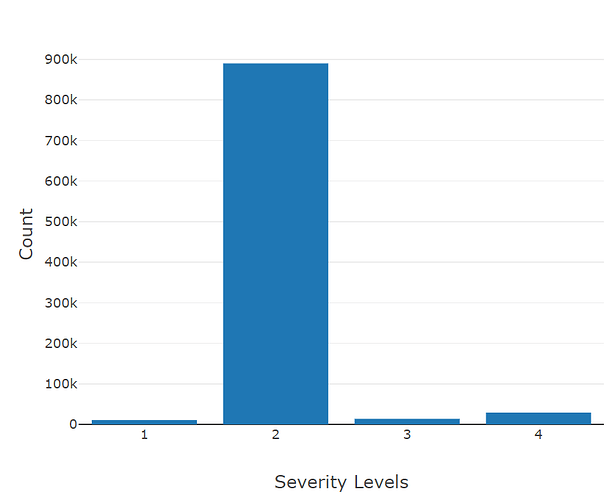
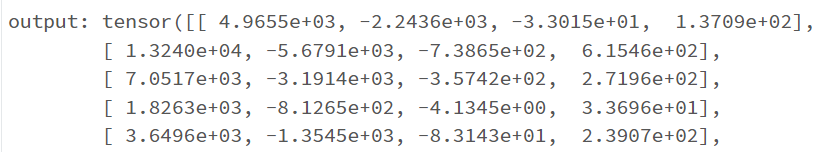Hi all,
I want to preface this by saying I’m relatively new to this field, so I apologize in advance if the solution is trivial. I’m working to build a prediction model that is able to take general information about the weather and location of an accident to predict the severity of traffic caused as a result of the crash. The dataset I’m using is from Kaggle (link to dataset).
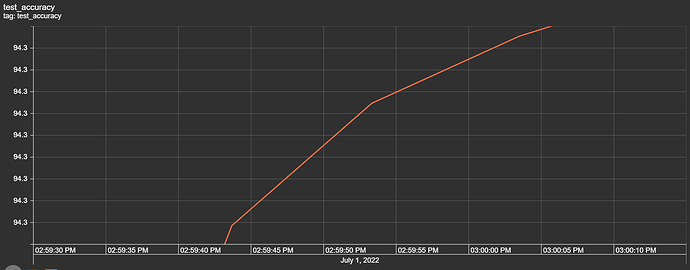
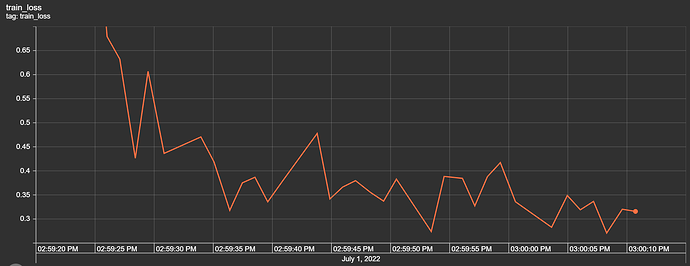
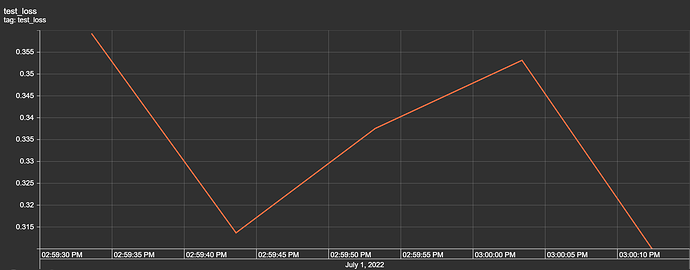
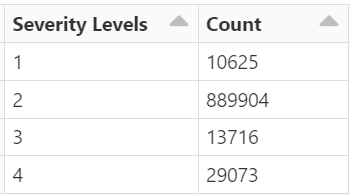
In particular, I’m attemping to use the fields (“Street”, “City”, “County”, “State”, “Zipcode”, “Temperature(F)”, “Wind_Chill(F)”, “Visibillity(mi)”, “Wind_Speed(mph)”, “Precipitation(in)”, and “Weather_Condition”) as the inputs (features) to the model to predict a severity value that ranges from 1 to 4 (labels).
All of the string fields (Street, City, County, State, Zipcode, and Weather_Condition) have been converted to pandas category types and encoded via “cat.codes”. All of the numerical values, with the exception of severity, have been converted to “np.float32” values. The severity is maintained as an integer value.
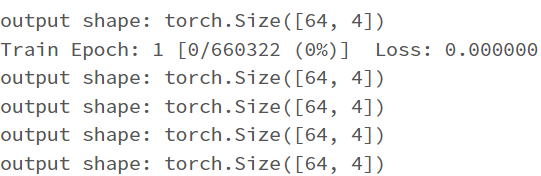
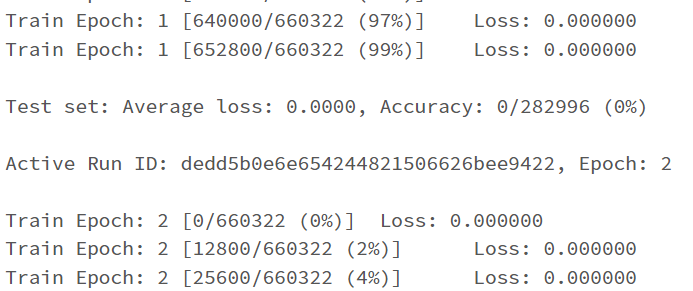
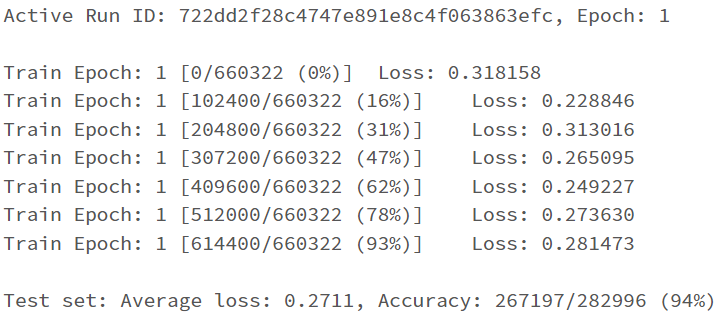
I split the dataset (train/test sets) via sklearn and run the following several commands to convert the training set into something iterable (DataLoaders):
#Convert Training/Test Datasets from a numpy.ndarray to a tensor.
X_train = torch.from_numpy(X_train)
y_train = torch.from_numpy(y_train)
X_train,y_train=X_train.type(torch.FloatTensor),y_train.type(torch.FloatTensor)
# y_train = torch.from_numpy(y_train).view(-1,1)
X_test = torch.from_numpy(X_test)
# y_test = torch.from_numpy(y_test).view(-1,1)
y_test = torch.from_numpy(y_test)
X_test,y_test=X_test.type(torch.FloatTensor),y_test.type(torch.FloatTensor)
# Make Tensor Data Iterable for Model Training
train = torch.utils.data.TensorDataset(X_train,y_train)
test = torch.utils.data.TensorDataset(X_test,y_test)
train_loader = torch.utils.data.DataLoader(train, batch_size = args.batch_size, shuffle = True, **kwargs)
test_loader = torch.utils.data.DataLoader(test, batch_size = args.test_batch_size, shuffle = True, **kwargs)
For my NN class definition, I do the following:
class ANN(nn.Module):
def __init__(self, input_dim = 11, output_dim = 4):
super(ANN, self).__init__()
self.fc1 = nn.Linear(input_dim, 64)
self.fc2 = nn.Linear(64, 64)
self.fc3 = nn.Linear(64, 32)
self.fc4 = nn.Linear(32, 32)
self.output_layer = nn.Linear(32,output_dim)
self.dropout = nn.Dropout(0.15)
def forward(self, x):
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.dropout(x)
x = F.relu(self.fc3(x))
x = F.relu(self.fc4(x))
x = self.output_layer(x)
return F.log_softmax(x, dim=1)
I have the first layer fc1 with the shape of (11,64) with the intention that I have 11 features/attributes being passed as inputs for the model. My goal is to have the model predict a distinct severity level (so a prediction should simply be 1, 2, 3, or 4; as severity levels are mutually exclusive). I thought as a result of this, the output layer needs to be (32,4) for the four different values of severity. However, when I attempt to train the model with this output layer size, I get a nan loss and the following warning: