Hello,
I have a dataset repesented as Tensor of shape (x, y), where x are observations and y are features (y is around 5k). What I want to achieve is to have first layer where pairs of input features are connected to each neuron separately, as in this image:
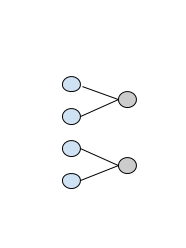
assuming the first layer (blue) is the input data and the second layer (gray) is the first hidden layer.
I would be able to follow the approach from Linear layer with custom connectivity (at least a similar one), but the problem is that I wanted to have ~2500 such neurons (or in another words, parallel/individual layers).
What I’ve come up with so far is the following (simplified case, but explains what I mean):
class CustomConnectivity(nn.Module):
def __init__(self):
self.indexer = [Variable(torch.LongTensor([i, i+1])) for i in range(0, 5000, 2)]
self.linear_list = nn.ModuleList([nn.Linear(2, 1) for _ in range(2500)])
self.fc_last = nn.Linear(2500, 1)
def forward(self, x):
lin_pre_outputs = [F.relu(self.linear_list[i](x.index_select(dim=1, index=self.indexer[i]))) for i in range(2500)]
x = torch.cat(lin_pre_outputs, dim=1)
x = F.sigmoid(self.fc_last(x))
return x
This is the fastest approach I could come up with, but unfortunately it does not run fast at all ![]() . The approaches with similar number of parameters seem to run 20-30x faster than this. This network is actually faster to train on CPU than on GPU, which I guess is the problem of input indexing and/or the for loop.
. The approaches with similar number of parameters seem to run 20-30x faster than this. This network is actually faster to train on CPU than on GPU, which I guess is the problem of input indexing and/or the for loop.
I tried the approach with creating fully connected later with zero’d weights and gradients, but despite of being much much uglier, it doesn’t really run faster and is much more memory consuming. I failed to implement some kind of sparse tensor approach (the API is undocumented).
What do you think is the most efficient (mostly computationally efficient, memory efficiency is less important) way to implement such network?