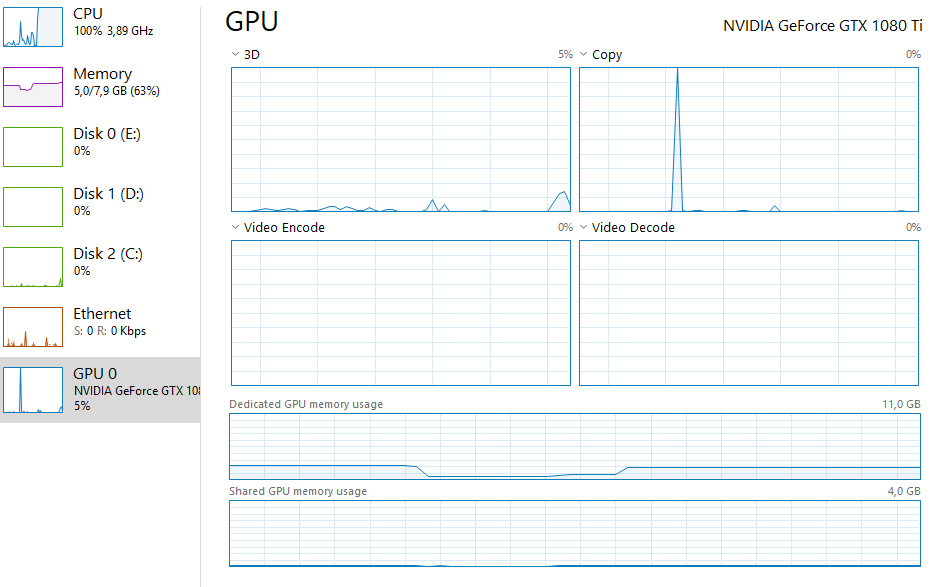
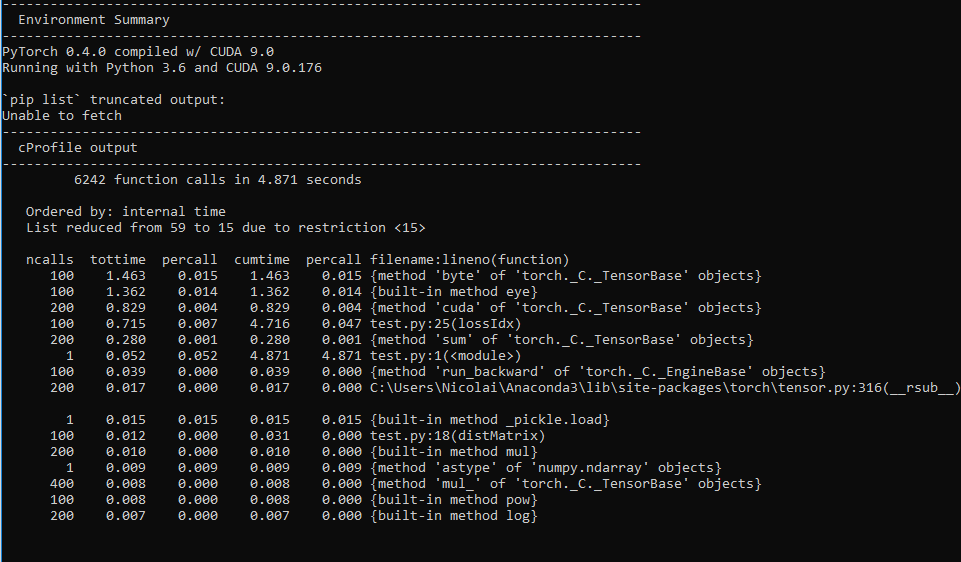
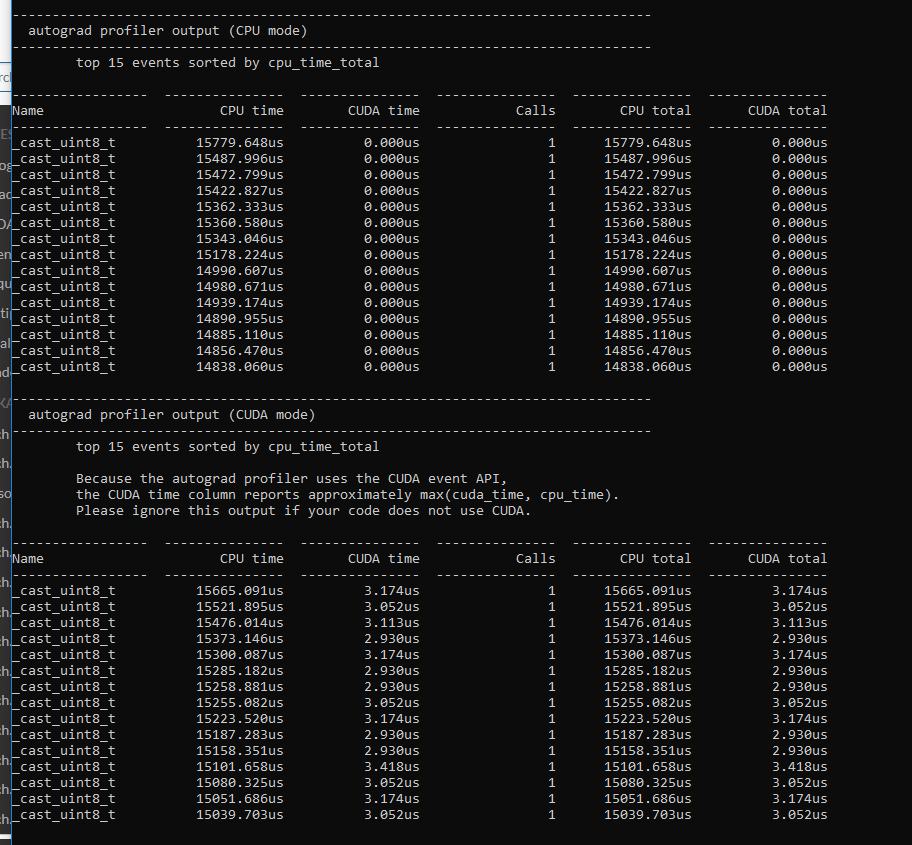
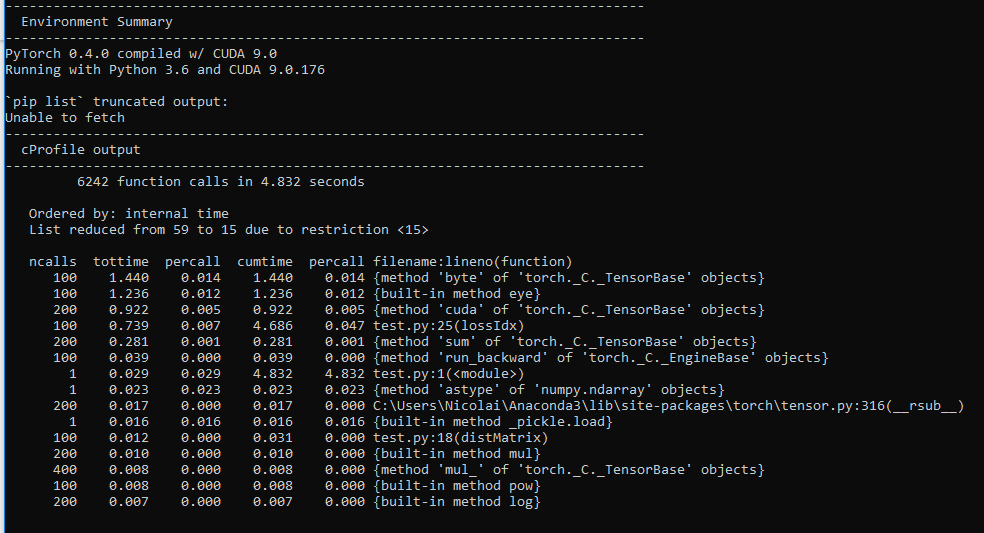
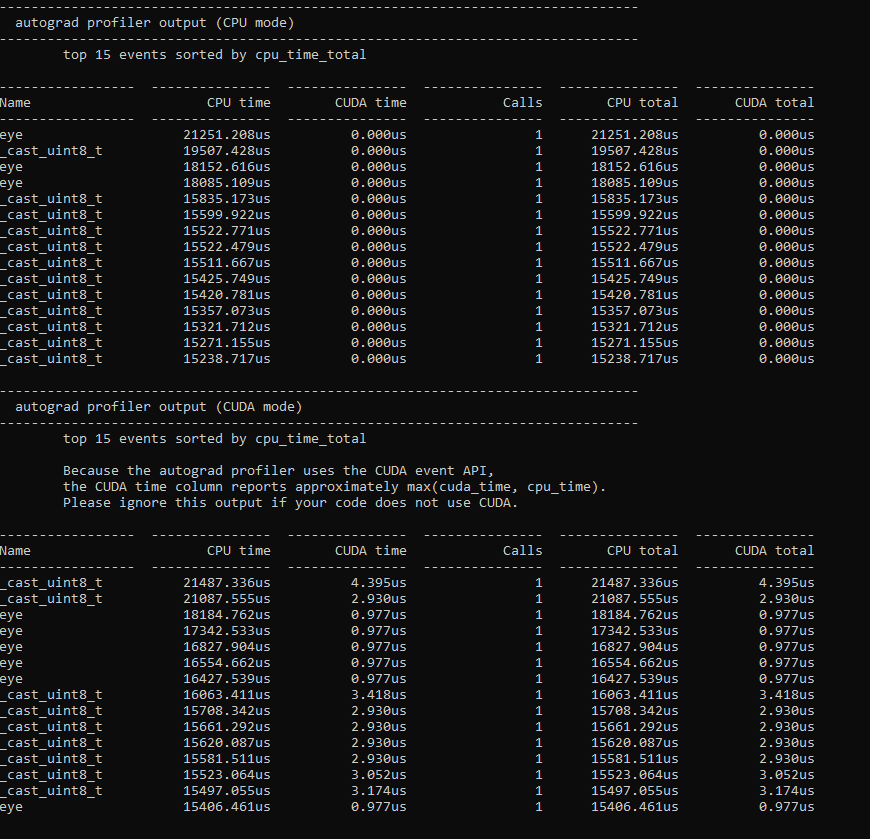
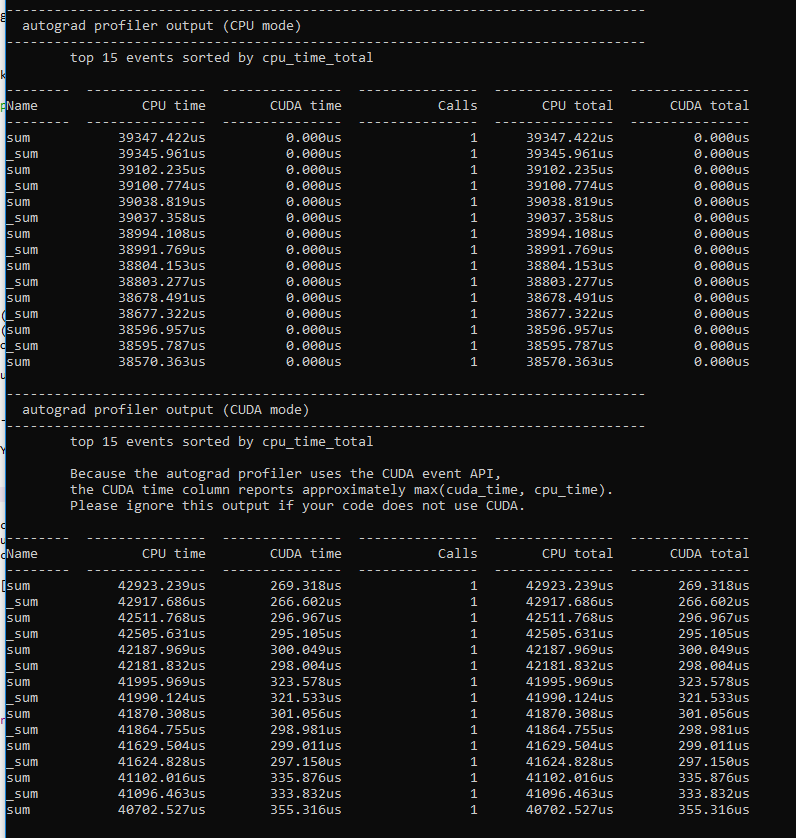
I wrote a model for doing latent space representation of graphs in PyTorch utilizing the autograd to minimize a cost function, this should according to my understanding be possible to run it on the GPU, but when I run my script i get 0% GPU utliziation.
import numpy as np
import torch
import torch.autograd as autograd
import pickle
import networkx as nx
'''
Pairwise euclidean distance
'''
def distMatrix(m):
n = m.size(0)
d = m.size(1)
x = m.unsqueeze(1).expand(n, n, d)
y = m.unsqueeze(0).expand(n, n, d)
return torch.sqrt(torch.pow(x - y, 2).sum(2) + 1e-4)
def lossIdx(tY):
d = -distMatrix(tZ)+B
sigmoidD = torch.sigmoid(d)
#calculating cost
reduce = torch.add(torch.mul(tY, torch.log(sigmoidD)), torch.mul((1-tY), torch.log(1-sigmoidD)))
#remove diagonal
reduce[torch.eye(n).byte().cuda()] = 0
return -reduce.sum()
def createBAnetwork(n, m, clusters):
a = nx.barabasi_albert_graph(n,m)
b = nx.barabasi_albert_graph(n,m)
c = nx.union(a,b,rename=('a-', 'b-'))
c.add_edge('a-0', 'b-0')
for i in range(clusters-2):
c = nx.convert_node_labels_to_integers(c)
c = nx.union(a,c,rename=('a-', 'b-'))
c.add_edge('a-0', 'b-0')
return(c)
c = createBAnetwork(1000,3,3)
Y = np.asarray(nx.adj_matrix(c).todense())
#loading adjacency matrix from pickle file
#Y = pickle.load(open( "data.p", "rb" ))
n = np.shape(Y)[0]
k = 2
Z = np.random.rand(n,k)
tZ = autograd.Variable(torch.cuda.FloatTensor(Z), requires_grad=True)
B = autograd.Variable(torch.cuda.FloatTensor([0]), requires_grad=True)
tY = autograd.Variable(torch.cuda.FloatTensor(Y.astype("uint8")), requires_grad=False)
optimizer = torch.optim.Adam([tZ, B], lr = 1e-3)
steps = 50000
for i in range(steps):
optimizer.zero_grad()
l = lossIdx(tY).cuda()
l.backward(retain_graph=True)
optimizer.step()
del l
It works, and it is solving the problem, just not on the GPU, but on the CPU.
Any help on whats going wrong or how to get this working on the GPU would be highly appreciated.
The file can be found here: https://drive.google.com/open?id=1j6kkD9-YNW11iFfVYBzjYrvssFNcqSXf