Hi everyone,
I have a question regarding the understanding of num_workers, I couldn’t answer it myself.
I have a data set that contains image data and the corresponding masks (segmentation task). The file format is HDF5.
I wrote a custom data set class that loads the data and use a data loader for loading the data in batches.
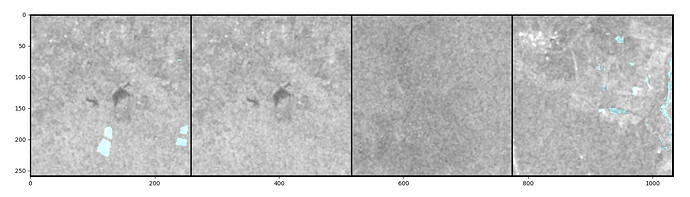
To visualize the data, I wrote a function that grids the image data and the corresponding masks and overlays the two, so that I can see the masks on the image. For that, I directly pass a batch coming from the dataloader to my “gridding function”.
As long as I use num_workers = 0 or 1, everything works as expected.
For num_workers > 1, it seems like my training samples and the corresponding masks are shuffled around (some images appear in several batches but with different masks, sometimes there’s no image at all, just the mask). I uploaded an example were you can see that the first two samples in the batch are identical but the mask (blueish parts) are different (and both wrong). This never happens if I set num_workers to 0 or 1.
I read that there are some problems with HDF5 datasets and multitasking, but I’m not sure if my problem relates to this or if there is something else going on that I’m unaware of.
Tech specs:
- Ubuntu 18.04
- i7-4720HQ CPU
- 16 GB RAM
- no CUDA, just CPU (for now)
Thank you for your help!