Hi everyone,
I have a directory with numpy files, each containing a data instance (for now, this image is generated randomly, without post processing - in my real scenario, it will be a numpy structured array, with some preprocessing).
I have implemented a map-style pytorch dataset, which loads the right numpy file every time getitem is called.
To avoid having to use shuffle=True (which shuffles the iterator), I have permuted the indices and saved them as an attribute. This had no effect on the performances.
I have created different dataloaders with various arguments passed (num_workers, batch_size, and etc.), and I have measured the performance across 30 batches, trying to see how the number of iterations/second changes with different parameters.
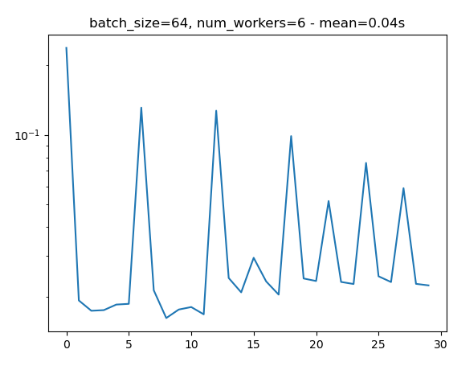
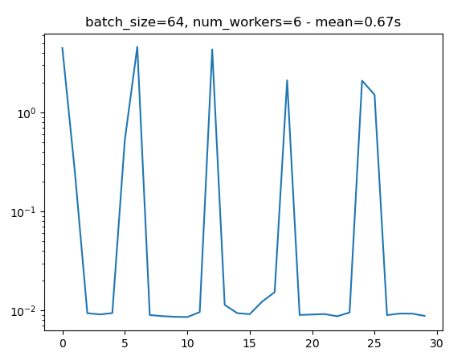
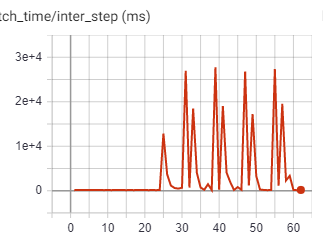
The two weird phenomena I see are:
- Inconsistency: Sometimes, the exact code runs significantly (x17) faster on the 2nd run than on the 1st run. This is terribly weird, and I have no idea what causes this.
- Periodical slowing: I noticed that when using num_workers=x, every xth iteration is about 10 times as slow as previous ones.
I would love it if someone could tell me why I see this behaviour and what I can do to overcome it.
Attached is a minimal reproducible example and 2 figures that show the weird behaviours (notice that these two runs have periodical nature, and both are run with the same parameters).
import time
import matplotlib.pyplot as plt
import numpy as np
import torch
from pathlib import Path
from tqdm import tqdm
from torch.utils.data import Dataset, DataLoader
root = Path('/home/yonatan/Desktop/NumpyDataset/data')
num_files = 6400
def generate_dataset(root, num_files):
for i in tqdm(range(N)):
mat = np.random.randint(low=0, high=255, size=(30, 300, 300))
mat = mat.astype('int8')
file_path = root / f'file_{i}.npy'
np.save(file_path, mat)
class NumpyDataset(Dataset):
def __init__(self, root, indxs):
self.root = root
self.indices = list(indxs)
np.random.seed(0)
self.permuted_indices = np.random.permutation(self.indices)
def __len__(self):
return len(self.indices)
def __getitem__(self, idx):
permuted_index = self.permuted_indices[idx]
return np.load(self.root / f'file_{permuted_index}.npy')
def measure_timing(N, batchsize, num_workers):
numpy_dataset = NumpyDataset(root=root, indxs=range(num_files))
numpy_dataloader = DataLoader(numpy_dataset,
batch_size=batchsize,
shuffle=False,
num_workers=num_workers,
pin_memory=False)
numpy_iter = iter(numpy_dataloader)
timings = [time.time()]
pbar = tqdm(total=N)
for i in range(N):
batch = next(numpy_iter)
timings.append(time.time())
pbar.update(1)
return np.diff(timings)
def measure_timing_and_plot_results(N, batch_size, num_workers):
timings = measure_timing(N=N, batchsize=batch_size, num_workers=num_workers)
total_mean = np.mean(timings)
plt.title(f'batch_size={batch_size}, num_workers={num_workers} - mean='
f'{total_mean:.2f}s')
plt.plot(timings)
plt.yscale('log')
plt.show()
if __name__ == '__main__':
# generate_dataset(root=root)
measure_timing_and_plot_results(N=30, batch_size=128, num_workers=6)