Dear all,
I tried to implement the " Optimistic mirror descent in saddle-point problems: Going the extra(-gradient) mile" algorithm, from here, built on top of the Adam optimizer.
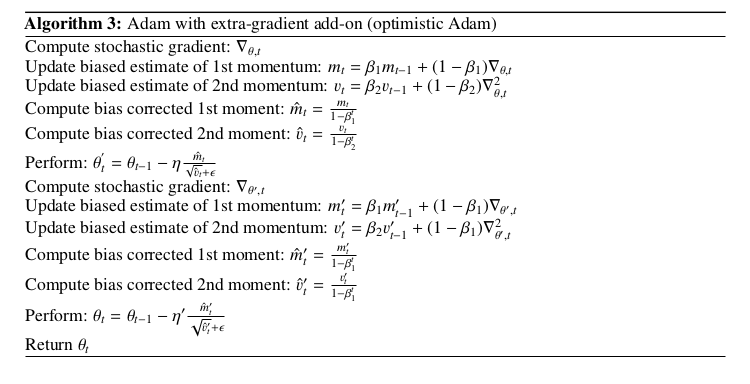
This is the algorithmic structure, and I am using common learning rate for both steps:
Could it please be possible for experts with pytorch to give me some feedback for my code (am newbie on pytorch)? This is the code:
import math
import torch
from torch.optim import Optimizer
class OptMirrorAdam(Optimizer):
"""Implements Optimistic Adam algorithm. Built on official implementation of Adam by pytorch.
See "Optimistic Mirror Descent in Saddle-Point Problems: Gointh the Extra (-Gradient) Mile"
double blind review, paper: https://openreview.net/pdf?id=Bkg8jjC9KQ
Standard Adam
It has been proposed in `Adam: A Method for Stochastic Optimization`_.
Arguments:
params (iterable): iterable of parameters to optimize or dicts defining
parameter groups
lr (float, optional): learning rate (default: 1e-3)
betas (Tuple[float, float], optional): coefficients used for computing
running averages of gradient and its square (default: (0.9, 0.999))
eps (float, optional): term added to the denominator to improve
numerical stability (default: 1e-8)
weight_decay (float, optional): weight decay (L2 penalty) (default: 0)
amsgrad (boolean, optional): whether to use the AMSGrad variant of this
algorithm from the paper `On the Convergence of Adam and Beyond`_
.. _Adam\: A Method for Stochastic Optimization:
https://arxiv.org/abs/1412.6980
.. _On the Convergence of Adam and Beyond:
https://openreview.net/forum?id=ryQu7f-RZ
"""
def __init__(self, params, lr=1e-3, betas=(0.9, 0.999), eps=1e-8,
weight_decay=0, amsgrad=False):
if not 0.0 <= lr:
raise ValueError("Invalid learning rate: {}".format(lr))
if not 0.0 <= eps:
raise ValueError("Invalid epsilon value: {}".format(eps))
if not 0.0 <= betas[0] < 1.0:
raise ValueError("Invalid beta parameter at index 0: {}".format(betas[0]))
if not 0.0 <= betas[1] < 1.0:
raise ValueError("Invalid beta parameter at index 1: {}".format(betas[1]))
defaults = dict(lr=lr, betas=betas, eps=eps,
weight_decay=weight_decay, amsgrad=amsgrad)
super(OptMirrorAdam, self).__init__(params, defaults)
def __setstate__(self, state):
super(OptMirrorAdam, self).__setstate__(state)
for group in self.param_groups:
group.setdefault('amsgrad', False)
def step(self, closure=None):
"""Performs a single optimization step.
Arguments:
closure (callable, optional): A closure that reevaluates the model
and returns the loss.
"""
loss = None
# Do not allow training with out closure
if closure is None:
raise ValueError("This algorithm requires a closure definition for the evaluation of the intermediate gradient")
# Create a copy of the initial parameters
param_groups_copy = self.param_groups.copy()
# ############### First update of gradients ############################################
# ######################################################################################
for group in self.param_groups:
for p in group['params']:
if p.grad is None:
continue
grad = p.grad.data
if grad.is_sparse:
raise RuntimeError('Adam does not support sparse gradients, please consider SparseAdam instead')
amsgrad = group['amsgrad']
state = self.state[p]
# @@@@@@@@@@@@@@@ State initialization @@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@
if len(state) == 0:
state['step'] = 0
# Exponential moving average of gradient values
state['exp_avg_1'] = torch.zeros_like(p.data)
state['exp_avg_2'] = torch.zeros_like(p.data)
# Exponential moving average of squared gradient values
state['exp_avg_sq_1'] = torch.zeros_like(p.data)
state['exp_avg_sq_2'] = torch.zeros_like(p.data)
if amsgrad:
# Maintains max of all exp. moving avg. of sq. grad. values
state['max_exp_avg_sq_1'] = torch.zeros_like(p.data)
state['max_exp_avg_sq_2'] = torch.zeros_like(p.data)
# @@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@
exp_avg1, exp_avg_sq1 = state['exp_avg_1'], state['exp_avg_sq_1']
if amsgrad:
max_exp_avg_sq1 = state['max_exp_avg_sq_1']
beta1, beta2 = group['betas']
# @@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@
# Step will be updated once
state['step'] += 1
# @@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@
if group['weight_decay'] != 0:
grad = grad.add(group['weight_decay'], p.data)
# Decay the first and second moment running average coefficient
exp_avg1.mul_(beta1).add_(1 - beta1, grad)
exp_avg_sq1.mul_(beta2).addcmul_(1 - beta2, grad, grad)
bias_correction1 = 1 - beta1 ** state['step']
bias_correction2 = 1 - beta2 ** state['step']
# *****************************************************
# Additional steps, to get bias corrected running means
exp_avg1 = torch.div(exp_avg1, bias_correction1)
exp_avg_sq1 = torch.div(exp_avg_sq1, bias_correction2)
# *****************************************************
if amsgrad:
# Maintains the maximum of all 2nd moment running avg. till now
torch.max(max_exp_avg_sq1, exp_avg_sq1, out=max_exp_avg_sq1)
# Use the max. for normalizing running avg. of gradient
denom1 = max_exp_avg_sq1.sqrt().add_(group['eps'])
else:
denom1 = exp_avg_sq1.sqrt().add_(group['eps'])
step_size1 = group['lr'] * math.sqrt(bias_correction2) / bias_correction1
p.data.addcdiv_(-step_size1, exp_avg1, denom1)
# Perform additional backward step to calculate stochastic gradient - WATING STATE
loss = closure()
#
# ############### Second evaluation of gradient step #######################################
# ######################################################################################
for (group, group_copy) in zip(self.param_groups,param_groups_copy ):
for (p, p_copy) in zip(group['params'],group_copy['params']):
if p.grad is None:
continue
grad = p.grad.data
if grad.is_sparse:
raise RuntimeError('Adam does not support sparse gradients, please consider SparseAdam instead')
amsgrad = group['amsgrad']
state = self.state[p]
exp_avg2, exp_avg_sq2 = state['exp_avg_2'], state['exp_avg_sq_2']
if amsgrad:
max_exp_avg_sq2 = state['max_exp_avg_sq_2']
beta1, beta2 = group['betas']
if group['weight_decay'] != 0:
grad = grad.add(group['weight_decay'], p.data)
# Decay the first and second moment running average coefficient
exp_avg2.mul_(beta1).add_(1 - beta1, grad)
exp_avg_sq2.mul_(beta2).addcmul_(1 - beta2, grad, grad)
# *****************************************************
# Additional steps, to get bias corrected running means
exp_avg2 = torch.div(exp_avg2, bias_correction1)
exp_avg_sq2 = torch.div(exp_avg_sq2, bias_correction2)
# *****************************************************
if amsgrad:
# Maintains the maximum of all 2nd moment running avg. till now
torch.max(max_exp_avg_sq2, exp_avg_sq2, out=max_exp_avg_sq2)
# Use the max. for normalizing running avg. of gradient
denom2 = max_exp_avg_sq2.sqrt().add_(group['eps'])
else:
denom2 = exp_avg_sq2.sqrt().add_(group['eps'])
step_size2 = group['lr'] * math.sqrt(bias_correction2) / bias_correction1
p_copy.data.addcdiv_(-step_size2, exp_avg2, denom2)
p = p_copy # pass parameters to the initial weight variables.
return loss
Thank you very much for your time.