Hi guys, i am working with a small dataset of Renal Ultrasound images (1985 images). In this case i am using a pretrained resnet152, fine tuning and data augmentation to do a binary classification between healthy and pathological kidneys. In addition, the dataset is unbalanced and there is 1400 images with the pathological label and the rest for healthy kidneys.
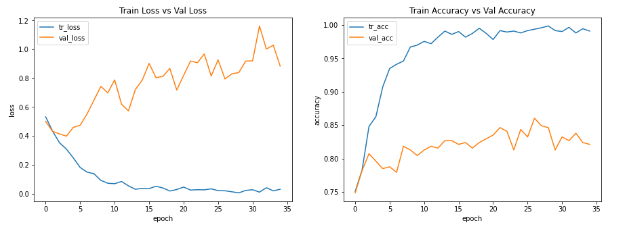
The problem is that when i try to get good results, i have overfitting and i don’t know what else to do. This is my code:
import random
from natsort import natsorted, ns
import skimage.io as io
import scipy.io as sio
from torchvision.transforms import Compose, ToTensor, Resize , Grayscale, RandomHorizontalFlip,RandomRotation,Normalize,CenterCrop
from ClaseEcografia import *
from BuclesTrValTest import *
import matplotlib.pyplot as plt
import torch
from torch.utils.data import DataLoader
from torchvision import datasets, transforms
import os
from time import time
from tqdm import tqdm
import numpy
from torch.nn import Linear, CrossEntropyLoss
from torch.optim import Adam
import torchvision
from torchvision.models import resnet152,ResNet152_Weights
import torch.nn.functional as F
import torch.nn as nn
import numpy as np
Transformacion_compuesta = Compose([ #[1]
Resize(256), #[2]
CenterCrop(224), #[3]
ToTensor(), #[4]
Normalize( #[5]
mean=[0.485, 0.456, 0.406], #[6]
std=[0.229, 0.224, 0.225] #[7]
)])
Transformacion_compuesta_train = Compose([ #[1]
Resize(256), #[2]
CenterCrop(224), #[3]
RandomHorizontalFlip(p=0.5),
RandomRotation(degrees=10),
ToTensor(), #[4]
Normalize( #[5]
mean=[0.485, 0.456, 0.406], #[6]
std=[0.229, 0.224, 0.225] #[7]
)])
root = ‘bbdd_kidney’ #Esta es la ruta relativa para nuestra base de datos
trainDataset = Ecografias(root, mode=‘train’,transform=Transformacion_compuesta_train)
testDataset = Ecografias(root, mode=‘test’,transform=Transformacion_compuesta)
validationDataset = Ecografias(root, mode=‘validation’,transform=Transformacion_compuesta)
print(“-------------------\n”)
print(“La longitud del conjunto de ‘Train’ es de " + str(len(trainDataset))+”\n")
print(“-------------------\n”)
print(“La longitud del conjunto de ‘Validation’ es de " + str(len(validationDataset))+”\n")
print(“-------------------\n”)
print(“La longitud del conjunto de ‘Test’ es de " + str(len(testDataset))+”\n")
print(“-------------------\n”)
device = ‘cuda’ if torch.cuda.is_available() else ‘cpu’
print(f’Using {device} device’)
train_dataloader = DataLoader(trainDataset, batch_size = 32, shuffle=True)
validation_dataloader = DataLoader(validationDataset, batch_size = 32, shuffle=False)
test_loader = DataLoader(testDataset, batch_size=32, shuffle = False)
model = resnet152(weights=ResNet152_Weights.DEFAULT)
for param in model.parameters():
param.requires_grad = True
model.fc = Linear(in_features=2048, out_features=2)
model = model.to(device)
for name, param in model.named_parameters():
print('Name: ', name, "Requires_Grad: ", param.requires_grad)
optimizer = Adam(model.parameters(), lr=1e-4)
loss_fn = CrossEntropyLoss()
epochs = 35
tr_loss = np.zeros(epochs)
val_loss = np.zeros(epochs)
tr_acc = np.zeros(epochs)
val_acc = np.zeros(epochs)
best_val_loss = 10e10
best_model = model
for t in range(epochs):
print(f"Epoch {t+1}\n----------------")
avg_tr_loss_e, tr_acc_e=train_loop_return(train_dataloader, model ,loss_fn, optimizer)
#avg_tr_loss_e, tr_acc_e=train_loop_return_scheduler(train_dataloader, model ,loss_fn, optimizer,scheduler)
#avg_train_loss_e, train_acc_e,train_prob_mask =validation_loop_return(train_loader, model ,loss_fn)
avg_val_loss_e, val_acc_e,val_prob_mask =validation_loop_return(validation_dataloader, model ,loss_fn)
#break;
if (avg_val_loss_e < best_val_loss):
best_model = model #Comprobar
tr_loss[t]= avg_tr_loss_e
val_loss[t]= avg_val_loss_e
tr_acc[t] =tr_acc_e
val_acc[t] =val_acc_e
print(“Done!”)
import matplotlib.pyplot as plt
fig = plt.figure(1, figsize=(15,5))
plt.subplot(1,2,1)
plt.plot(range(epochs), tr_loss)
plt.plot(range(epochs), val_loss)
plt.xlabel(‘epoch’)
plt.ylabel(‘loss’)
plt.legend([‘tr_loss’, ‘val_loss’])
plt.title(‘Train Loss vs Val Loss’)
plt.subplot(1,2,2)
plt.plot(range(epochs), tr_acc)
plt.plot(range(epochs), val_acc)
plt.xlabel(‘epoch’)
plt.ylabel(‘accuracy’)
plt.legend([‘tr_acc’, ‘val_acc’])
plt.title(‘Train Accuracy vs Val Accuracy’)
def train_loop_return(dataloader, model, loss_fn, optimizer):
size = len(dataloader.dataset)
num_batches = len(dataloader)
train_loss, correct = 0,0
for batch, (X,y) in enumerate(dataloader):
#Movemos los datos a la CPU para acelerar el proceso
X = X.to(device)
y = y.to(device)
# print(y)
#Calculamos la predicción y la pérdida
pred = model(X)
loss = loss_fn(pred,y)
# Propagación hacia atrás
optimizer.zero_grad() #Inicializa a cero el valor de los gradientes de los parámetros del modelo
loss.backward() #Propagación el error de la función de pérdida para estimar el gradiente de cada parámetro con respecto del error de predicción
optimizer.step() #Una vez tenemos nuestros gradientes llamamos a optimizer.step() para ajustar los parámetros con los gradientes calculados en el paso anterior.
train_loss += loss.item()
correct += (pred.argmax(1)==y).type(torch.float).sum().item()
if batch % 100 == 0: #Imprimimos cada 100 batches
loss, current = loss.item(), batch * len(X)
print(f"loss: {loss:>7f} [{current:>5d}/{size:>5d}]")
avg_train_loss = train_loss / num_batches
train_accuracy = correct/size
print(f"Train Error: \n Accuracy: {(100*train_accuracy):>0.1f}%, Avg loss: {avg_train_loss:>8f} \n")
return avg_train_loss, train_accuracy
def validation_loop_return(dataloader, model, loss_fn):
size = len(dataloader.dataset)
num_batches = len(dataloader)
validation_loss, correct = 0,0
bs = dataloader.batch_size
probs = torch.zeros(size,2)
with torch.no_grad():
for nb, (X,y) in enumerate(dataloader):
X = X.to(device)
y = y.to(device)
pred = model(X)
validation_loss +=loss_fn(pred,y).item()
correct += (pred.argmax(1) == y).type(torch.float).sum().item() #Va almacenando todas las muestras que han sido correctas.
pred_probab = nn.Softmax(dim=1)(pred)
probs[nb*bs:nb*bs+bs] = pred_probab
#print(nb)
avg_validation_loss = validation_loss / num_batches
validation_accuracy = correct/size
print(f"Validation Error: \n Accuracy: {(100*validation_accuracy):>0.1f}%, Avg loss: {avg_validation_loss:>8f} \n")
return avg_validation_loss, validation_accuracy, probs[:,1]
These are the graphic results for train and validation. I would really appreciate the help