Hi everyone,
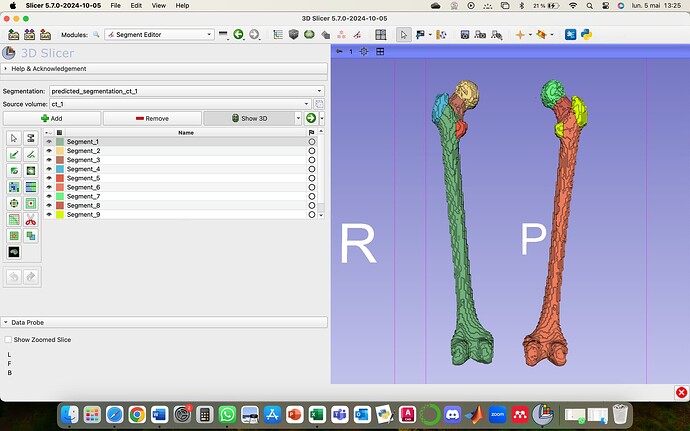
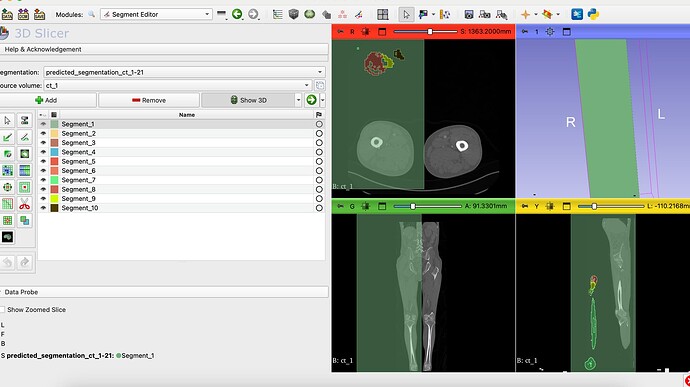
I want to create a view a prediction done by a training and validation Attention UNet code. I have some segmentations of femur. The segmentations contain 10 classes each, from 1 to 10. I used Cross Entropy and i preprocessed all the CT scans before everything else i. My Dices are great, all of them are up to 0.92 for training and validation, but when I try to view the prediction in 3D slicer, there are only 9 segments. I think the problem is that with the Cross Entropy we train classes from 0 to 9 and then the 0 class is forget.
Here is the part of the processing function in my Attention UNet code:
def prepare(self):
for ct_path, (left_path, right_path) in tqdm(self.ct_paths):
ct_volume = read_nii(ct_path)
label_volume = read_nii(left_path) # Already combined masks into 10 classes
for idx in range(ct_volume.shape[-1]):
img = cv2.resize(ct_volume[..., idx], (self.img_size, self.img_size), interpolation=cv2.INTER_AREA).astype('float32')
label = cv2.resize(label_volume[..., idx], (self.img_size, self.img_size), interpolation=cv2.INTER_NEAREST)
img = (img - img.min()) / (img.max() - img.min() + 1e-5)
label = np.clip(label, 0, NUM_CLASSES-1).astype(np.uint8)
self.data.append(img[..., np.newaxis])
self.labels.append(label)
And from the .pth loading:
def read_nii(path):
return np.rot90(np.array(nib.load(path).get_fdata()))
def preprocess_slice(slice_2d):
resized = cv2.resize(slice_2d, (IMG_SIZE, IMG_SIZE), interpolation=cv2.INTER_AREA).astype(‘float32’)
normed = (resized - resized.min()) / (resized.max() - resized.min() + 1e-5)
return normed
def predict_volume(volume):
original_shape = volume.shape
predictions =
for i in tqdm(range(volume.shape[-1]), desc=“Predicting slices”):
slice_2d = volume[…, i]
input_tensor = preprocess_slice(slice_2d)
input_tensor = torch.tensor(input_tensor).unsqueeze(0).unsqueeze(0).to(DEVICE)
with torch.no_grad():
output = model(input_tensor)
pred = torch.argmax(torch.softmax(output, dim=1), dim=1).squeeze().cpu().numpy()
pred_resized = cv2.resize(pred.astype(‘uint8’), (original_shape[0], original_shape[1]), interpolation=cv2.INTER_NEAREST)
predictions.append(pred_resized)
pred_volume = np.stack(predictions, axis=-1)
return pred_volume