Hello Pytorch!
I am new to pytorch, and I’m trying to translate my sklearn MLPRegressor model into pytorch. The in-sample R-squared is better than sklearn, however, the out-of-sample R-squared is horrible.
My database details are as follows:
- Sample size: ~60k
- Feature size: 52 (including binary features)
I already did standardization for the features.
MLPRegressor model structure is:
hyperparams = {
"hidden_layer_sizes": (64, 32, 16),
"activation": "relu",
"solver": "adam",
"learning_rate_init": 0.01,
"alpha": 0.01,
"early_stopping": True,
"batch_size": 500,
"max_iter": 1000,
}
MLP_regr = MLPRegressor(**hyperparams)
the in-sample R2 for this model is ~0.6 and out-of-sample R2 is 0.2.
My PyTorch MLP implementation structure is
class MLP(nn.Module):
def __init__(self, in_dim, out_dim=1):
super(MLP, self).__init__()
self.layers = nn.Sequential(
nn.Linear(in_dim, 64),
nn.ReLU(),
nn.Linear(64, 32),
nn.ReLU(),
nn.Linear(32, 16),
nn.ReLU(),
nn.Linear(16, 1)
)
def forward(self, x):
return self.layers(x)
and the training procedure is:
num_epochs = 1000
batch_size = 500
loss_func = nn.MSELoss()
optimizer = torch.optim.Adam(mlp_torch.parameters(), lr=0.01, weight_decay=0.01)
trainloader = DataLoader(train_dataset, batch_size=batch_size)
testloader = DataLoader(test_dataset, batch_size=batch_size)
mlp_torch = MLP(in_dim=X_train_torch.size(1))
### training step
in_sample_r2_ = []
for epoch in range(num_epochs):
in_sample_r2_temp = []
for id_batch, (X_batch, y_batch) in enumerate(trainloader):
optimizer.zero_grad()
y_pred = mlp_torch(X_batch)
loss = loss_func(y_pred, y_batch.unsqueeze(1))
loss.backward()
optimizer.step()
# store in-sample R-squared
in_sample_r2_temp.append(r2_score(y_batch.detach().numpy(), y_pred.T[0].detach().numpy()))
in_sample_r2_.append(np.mean(in_sample_r2_temp))
if epoch % 50 == 1:
print(f"Epoch {epoch}: {loss.item()}, averaged in-sample R-squared is: {np.mean(in_sample_r2_temp)}")
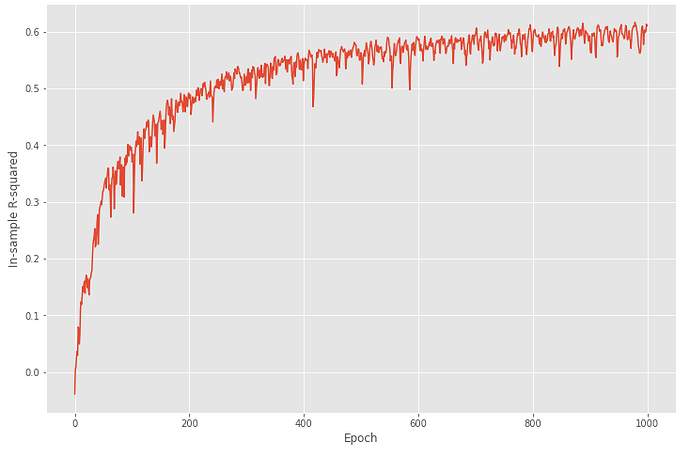
and the plot of in-sample R2 for my implementation looks like:
After 1000 epochs, my torch MLP achieves 0.61 in-sample R2, and according to the plot, I personally think it learns well. But when I test my model on the testing sets and calculate the out-of-sample R2, it surprisingly gives me a -0.6. Here is the code for testing step:
### testing step
with torch.no_grad():
out_sample_r2_ = []
for id_batch, (X_test, y_test) in enumerate(testloader):
y_pred = mlp_torch(X_test)
out_sample_r2 = r2_score(y_test.detach().numpy(), y_pred.T[0].detach().numpy())
out_sample_r2_.append(out_sample_r2)
print(f"Out-of sample R-squared: {np.mean(out_sample_r2_)}")
I am very confused about this result, it doesn’t seem like overfit and it has the similar result as sklearn does, but why the OOS R-squared too bad? Or did I make mistake to calculate the result? Thank you!!
Best,
-P