Greetings everyone,
I’m currently trying to implement a model originally developed with DeepTrack for particle tracking on video recordings. It’s built on top of Keras, so the model will use layers defined in Keras. The original model’s architecture is as follows (input image size is 64x64 but can be subject of changes):
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_1 (InputLayer) [(None, 64, 64, 1)] 0
conv2d (Conv2D) (None, 64, 64, 16) 160
activation (Activation) (None, 64, 64, 16) 0
max_pooling2d (MaxPooling2D (None, 32, 32, 16) 0
)
conv2d_1 (Conv2D) (None, 32, 32, 32) 4640
activation_1 (Activation) (None, 32, 32, 32) 0
max_pooling2d_1 (MaxPooling (None, 16, 16, 32) 0
2D)
conv2d_2 (Conv2D) (None, 16, 16, 64) 18496
activation_2 (Activation) (None, 16, 16, 64) 0
max_pooling2d_2 (MaxPooling (None, 8, 8, 64) 0
2D)
flatten (Flatten) (None, 4096) 0
dense (Dense) (None, 32) 131104
activation_3 (Activation) (None, 32) 0
dense_1 (Dense) (None, 32) 1056
activation_4 (Activation) (None, 32) 0
dense_2 (Dense) (None, 2) 66
=================================================================
Total params: 155,522
Trainable params: 155,522
Non-trainable params: 0
_________________________________________________________________
Note: The “Activation” layer is “ReLU”; convolution layers padding is set as “same”, kernel filter size is (3,3)
I generate synthetic data using DeepTrack data generator, which allows me to create images of particles recorded with a microscope. An example is the following:
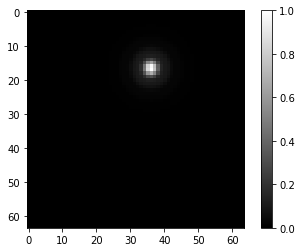
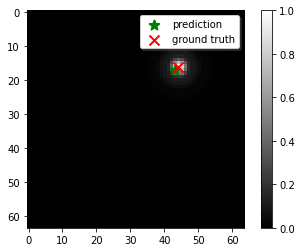
So the model is trained to find the center of these particles as labels. After training this model with a random generated dataset, I obtain some really good predictions on my validation sample. For example, the following image shows the accuracy level I achieve with this model:
Now my problem is that I want to translate this model to PyTorch (first) by loading the weights and biases of my Keras model into the PyTorch equivalent. After I verify that the PyTorch model works fine I will then move to Brevitas to quantize this model and then create an IP for an FPGA using the FINN compiler. The model I built is the following (it should be equivalent to the Keras one)
import torch.nn as nn
convolution_sizes = [16, 32, 64]
dense_sizes = [32, 32]
class SPTModel(nn.Module):
def __init__(self, in_channels: int, image_size: int, convolution_sizes: list, dense_sizes: list, output_size: int) -> None:
super(SPTModel, self).__init__()
self.layers = nn.Sequential()
in_size = in_channels
for idx, size in enumerate(convolution_sizes, 1):
self.layers.add_module(
name=f"conv_{idx}",
module=nn.Conv2d(in_size, size, kernel_size=(3, 3), padding="same")
)
self.layers.add_module(
name=f"conv_{idx}_relu",
module=nn.ReLU()
)
self.layers.add_module(
name=f"maxpool_{idx}",
module=nn.MaxPool2d((2, 2))
)
in_size = size
self.layers.add_module(name="flatten", module=nn.Flatten())
# the convolution layers squeeze the image of a factor 2
# after len(convolution_sizes) layers, the final size will be
# convolution_sizes[-1]*((image_size / (2**len(convolution_sizes)))**2)
maxpool_final_size = image_size // (2**len(convolution_sizes))
in_size = convolution_sizes[-1]*(maxpool_final_size*maxpool_final_size)
for idx, size in enumerate(dense_sizes, 1):
self.layers.add_module(
name=f"linear_{idx}",
module=nn.Linear(in_size, size)
)
self.layers.add_module(
name=f"linear_{idx}_relu",
module=nn.ReLU()
)
in_size = size
self.layers.add_module(
name=f"linear_output",
module=nn.Linear(dense_sizes[-1], output_size)
)
def forward(self, x):
x = self.layers(x)
return x
model_torch = SPTModel(in_channels=1, image_size=IMAGE_SIZE, convolution_sizes=convolution_sizes, dense_sizes=dense_sizes, output_size=2)
Initially I tried to train this same model using the same dataset, but the accuracy was poor everytime I tried. After some time I decided to give up and try to load the weights and biases of my Keras model into the PyTorch one as follows:
keras_weights = {w.name: w for w in model.weights}
torch_weights = model_torch.state_dict()
new_weights = {}
for k_layer, t_layer in zip(keras_weights.keys(), torch_weights.keys()):
if "conv" in k_layer and "kernel:0" in k_layer:
if "conv" in t_layer and "weight" in t_layer:
new_weights[t_layer] = torch.Tensor(np.moveaxis(keras_weights[k_layer], [-1, -2], [0, 1])).to(device)
elif "dense" in k_layer and "kernel:0" in k_layer:
if "linear" in t_layer and "weight" in t_layer:
new_weights[t_layer] = torch.Tensor(np.transpose(np.array(keras_weights[k_layer]))).to(device)
else:
new_weights[t_layer] = torch.Tensor(np.array(keras_weights[k_layer])).to(device)
model_torch.load_state_dict(new_weights)
After doing this, I tried again on my validation dataset to see if this somehow managed to fix the problem. Instead, what I find is that the performance is still poor:
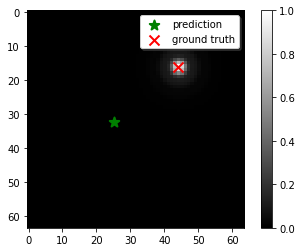
What’s going on here?
Some extra informations:
- loss function used with the Keras model is MSE
- optimizer used with Keras model is Adam (lr = 0.001)
- batch size: 1
- number of epochs: 128
- particle position in the image is randomly generated and the data generator provided by DeepTrack allows to continously generate new pseudo-random particles