I use some nets,FCN8 ,SegNet for semantic segmentation .The trouble follow:
all of the nets I used,The last layers of this net output the feature maps is (1,22,256,256),why not (1,3,256,256)? and another question is that the labels size is (1,1,256,256),why not(1,3,256,256)?
please help me, I a new gay
The output and label shape depend on your classification problem. If the last layer outputs an activation of [batch_size, 22, width, height], it probably means that the model was used to classify each pixel into one of 22 classes.
You should change the number of output channels to your use case.
Edit: I edited your title so that the question becomes clearer.
22 = 21 (classes) + 1 (background) in PASCAL_VOC dataset.
Thank your for you help . I know it by debug
Thank you for you help.Thank you very much .I know it
I am sorry,I want to ask for you another question.The label size is (batch_size, 1, width, height),that means the lable have one channel,so it is a picture?
And,the output size is (batch_size, class_num, width, height),then the cost function to compute loss by the every channel of the output and the only channel of the label?
Yes, the label seems to be a picture. You could try to visualize it using something like:
import matplotlib.pyplot as plt
plt.imshow(label[0, 0, ...].data.numpy())
When you are dealing with a multi-class problem, you could use NLLLoss or CrossEntropyLoss. Have a look at the doc.
Here is a small example with 22 classes and a binary example:
# Multi-class example
x = Variable(torch.randn(1, 22, 10, 10))
y = Variable(torch.LongTensor(1, 10, 10).random_(22))
criterion = nn.NLLLoss()
loss = criterion(x, y)
# Binary example
x = F.sigmoid(Variable(torch.randn(1, 1, 10, 10)))
y = Variable(torch.FloatTensor(1, 1, 10, 10).random_(2))
criterion = nn.BCELoss()
loss = criterion(x, y)
Thank you for you help ,good friend
I am facing similar problems. What should be input dimension in case of multi-class labels. I have image with 3 channels and labels with 1 channel and 12 classes.
I get output prediction with 12 channels. How do I tackle this ?
Thanks in advance!
If you are dealing with a multi-class classification, you could use nn.CrossEntropyLoss with a model output of [batch_size, nb_classes, h, w] and a target of [batch_size, h, w] containing the class indices.
How is your current target image defined?
Thank you for your reply.
Sorry for incomplete information
This is a semantic segmentation problem
Input data: Image is in .jpg format. I use transform.ToTensor() to convert this.
Target: Annotated .png image with 12 classes. I convert it using “torch.from_numpy(np.array(mask)).long()”
Following is the code I am working on.
data loader class
class GetDataset(torch.utils.data.Dataset):
def init(self, root, transformation=None):
self.root = root
self.transformation = transformation
# load all image files, sorting them to
# ensure that they are aligned
self.imgs = list(sorted(os.listdir(os.path.join(root, “JPEGImages_test”))))
self.masks = list(sorted(os.listdir(os.path.join(root, “SegmentationClass_test”))))
def __getitem__(self, idx):
# load images ad masks
img_path = os.path.join(self.root, "JPEGImages_test", self.imgs[idx])
mask_path = os.path.join(self.root, "SegmentationClass_test", self.masks[idx])
print(idx)
#import pdb
#pdb.set_trace()
img = Image.open(img_path)
mask = Image.open(mask_path)
img_t = self.transformation(img)
mask_t = torch.from_numpy(np.array(mask)).long()
return img_t, mask_t
def __len__(self):
return len(self.imgs)
==> Tried both losses
#criterion = nn.CrossEntropyLoss()
criterion = nn.NLLLoss()
Train function
def train_model(model, criterion, optimizer, dataloaders, scheduler, num_epochs=1):
import copy
best_model_wts = copy.deepcopy(model.state_dict())
print(num_epochs)
for epoch in range(num_epochs):
print('Epoch {}/{}'.format(epoch, num_epochs - 1))
print('-' * 10)
model.train() # Set model to training mode
for inputs, labels in dataloaders:
print("reached")
inputs = inputs.to(device)
type(input)
labels = labels.to(device)
optimizer.zero_grad()
outputs = model(inputs)
#import pdb
#pdb.set_trace()
loss = criterion(outputs['out'], labels)
loss.backward()
optimizer.step()
best_model_wts = copy.deepcopy(model.state_dict())
# load best model weights
model.load_state_dict(best_model_wts)
return model
#Train model
model_ft = train_model(model_segdl, criterion, optimizer_ft,data_loader, exp_lr_scheduler, num_epochs=1)
model_ft.eval()
x = torch.rand(1, 3, 300, 400)
predictions = model_ft(x)
predictions[‘out’].size()
Out[59]: torch.Size([1, 12, 300, 400])
Is this correct ? If not what I am missing
Could you print the shape of mask_t?
How are your masks stored? Is each class corresponding to a certain color in an RGB image or do the image files already contain class indices?
Yes I have indices for each class. mask_t are essentially labels, when I load them from dataloader, the shape is as follows.
Although I used label inputs with dimension of [1, 512, 512](which is my .png file) and converted it to long using torch.from_numpy(np.array(mask)).long()
ipdb> labels.size()
torch.Size([3, 512, 512])
ipdb> labels
tensor([[[2, 2, 2, …, 2, 2, 2],
[2, 2, 2, …, 2, 2, 2],
[2, 2, 2, …, 2, 2, 2],
…,
[2, 2, 2, …, 2, 2, 2],
[2, 2, 2, …, 2, 2, 2],
[2, 2, 2, …, 2, 2, 2]],
[[2, 2, 2, ..., 2, 2, 2],
[2, 2, 2, ..., 2, 2, 2],
[2, 2, 2, ..., 2, 2, 2],
...,
[9, 9, 9, ..., 2, 2, 2],
[9, 9, 9, ..., 2, 2, 2],
[9, 9, 9, ..., 2, 2, 2]],
[[1, 1, 1, ..., 2, 2, 2],
[1, 1, 1, ..., 2, 2, 2],
[1, 1, 1, ..., 2, 2, 2],
...,
[1, 1, 1, ..., 2, 2, 2],
[1, 1, 1, ..., 2, 2, 2],
[1, 1, 1, ..., 2, 2, 2]]])Based on the example output, it looks like each channel contains different values.
Are you sure that the mask image does not contain a color code?
Could you try to call target.view(target.size(0), -1).unique(dim=1) and post the unique color values here?
I had converted image to grayscale, shall I use annotated .png image ?
I’m not sure what the annotated .png images are, but your image does not seem to be grayscale, as the channels contain different values.
What did the unique call return?
labels.view(labels.size(0), -1).unique(dim=1)
tensor([[ 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3, 3,
3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3,
3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 5, 5, 5, 5, 5, 5, 5,
5, 5, 5, 5, 5, 5, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6,
6, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 8, 8, 8, 8,
8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8,
8, 9, 9, 9, 9, 9, 9, 9, 9],
[ 2, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 4, 4, 4, 5,
5, 5, 5, 5, 6, 6, 6, 7, 7, 7, 7, 7, 8, 8, 8, 8, 8, 9,
9, 9, 9, 9, 9, 9, 9, 10, 10, 10, 10, 11, 11, 11, 11, 11, 2, 2,
2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 4, 6, 6, 7, 7, 7, 7,
8, 8, 8, 8, 9, 9, 9, 9, 9, 11, 11, 2, 2, 2, 3, 3, 3, 9,
9, 9, 10, 10, 11, 11, 2, 2, 2, 5, 6, 7, 7, 7, 9, 9, 10, 11,
11, 2, 2, 4, 4, 7, 7, 9, 9, 10, 10, 10, 11, 11, 2, 2, 2, 2,
2, 2, 3, 3, 3, 3, 3, 3, 6, 7, 7, 8, 8, 8, 8, 9, 9, 9,
9, 2, 2, 3, 3, 7, 7, 10, 10],
[ 1, 2, 3, 5, 6, 7, 8, 9, 2, 3, 6, 7, 8, 9, 2, 7, 9, 1,
2, 3, 8, 9, 1, 2, 6, 2, 5, 6, 7, 9, 2, 3, 6, 8, 9, 1,
2, 3, 5, 6, 7, 8, 9, 1, 2, 3, 5, 2, 3, 5, 6, 7, 2, 3,
5, 6, 7, 8, 9, 2, 3, 6, 7, 8, 9, 7, 2, 6, 2, 5, 6, 7,
2, 3, 6, 9, 2, 3, 5, 6, 7, 3, 7, 2, 3, 8, 2, 3, 8, 2,
3, 8, 2, 3, 2, 3, 2, 5, 9, 2, 2, 2, 5, 9, 2, 5, 2, 2,
5, 2, 3, 2, 3, 2, 5, 2, 3, 2, 3, 5, 2, 3, 2, 3, 6, 7,
8, 9, 2, 3, 6, 7, 8, 9, 2, 2, 6, 2, 3, 6, 9, 2, 3, 6,
7, 2, 9, 2, 9, 2, 9, 2, 3]])
I definitely sense some problem here
It looks a bit strange.
I assumed your tensor has a shape of [channels, height, width], which should yield a clean output of the different color values.
Are you using an additional batch dimension?
If so, could you slice the tensor and run the code again?
Anyway, the values look a bit fishy and something might be wrong.
Could you also try to plot the image using e.g. matplotlib?
Thats a sample .png image
plt.imshow(img)
Out[10]: <matplotlib.image.AxesImage at 0x1288d4da0>
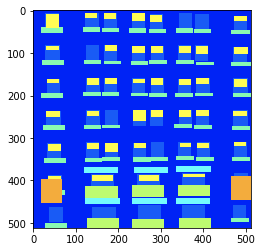
img.size
Out[12]: (512, 512)
np.unique(np.array(img))
Out[13]: array([2, 3, 5, 6, 7, 8, 9], dtype=uint8)
This looks perfectly fine.
Could you use this target image and try to recreate the loading logic from your Dataset and check, which method creates these pseudo-values?