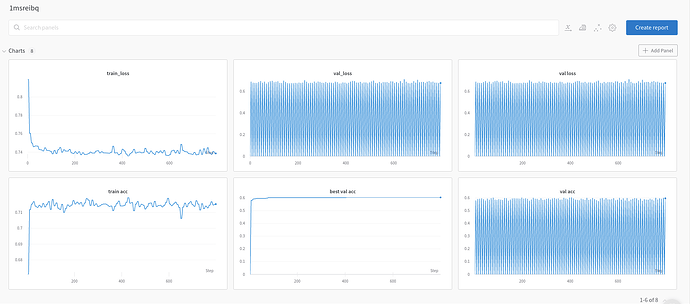
I am using an Inception V3 model pre-trained on ImageNet and when I train it on Bees and Ants dataset from PyTorch training, I get this result:
Dataset statistics:
* Train
* Bees: 121
* Ants: 124
* Val:
* Bees: 83
* Ants: 70
Best validation accuracy: 0.95 at epoch 6 (trained for 100 epochs)
However, when I try it on my own medical images, for also binary classification problem when everything apart from STD/mean vectors are the same, I get really poor results:
* Number of images of size 512x512 w 3 channels
* Train label +: 34,913
* Train label -: 27,785
* Val label +: 93,05
* Val label -: 5,940
Best val Acc: 0.609708 and best epoch 31
Could you please suggest some ways to look into this problem and try to debug it? What are some methods I could do to improve the accuracy of the validation set? Also, does PyTorch have any built-in tool to measure the domain gap or dissimilarity between ImageNet and each of these two datasets, namely my own in-house medical data and also bees/ants data?
I have used the following formula for calculating the STD and mean vectors for train/test/val and values are shown as below in the comment for my medical data:
data_transforms = {
'train': transforms.Compose([
transforms.RandomResizedCrop(input_size),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
#transforms.Normalize([0.7031, 0.5487, 0.6750], [0.2115, 0.2581, 0.1952])
]),
'val': transforms.Compose([
transforms.Resize(input_size),
transforms.CenterCrop(input_size),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
#transforms.Normalize([0.7016, 0.5549, 0.6784], [0.2099, 0.2583, 0.1998])
]),
'test': transforms.Compose([
transforms.Resize(input_size),
transforms.CenterCrop(input_size),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
#transforms.Normalize([0.7048, 0.5509, 0.6763], [0.2111, 0.2576, 0.1979])
])
}
# get the mean var std of train, test and val set for data transform
def get_mean_std(loader):
# VAR[X] = E[X**2] - E[X]**2
channels_sum, channels_squared_sum, num_batches = 0, 0, 0
for data, _ in loader:
channels_sum += torch.mean(data, dim=[0,2,3])
channels_squared_sum += torch.mean(data**2, dim=[0,2,3])
num_batches += 1
mean = channels_sum/num_batches
std = (channels_squared_sum/num_batches - mean**2)**0.5
return mean, std