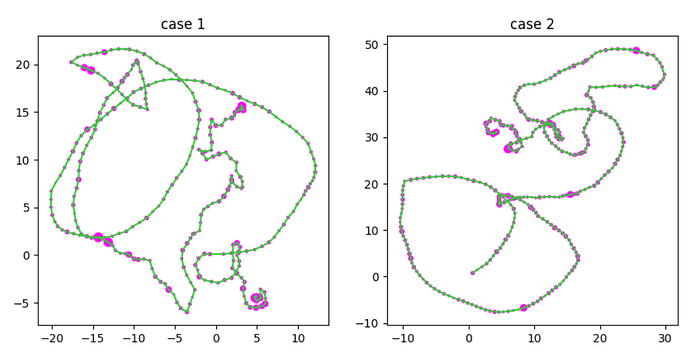
Hi everyone, My name is Josh and I’m quite new to using GNNs (and machine learning in general). I have a dataset that consists of 4 columns, 3 of which pertain to the position of an object (x,y, and z coordinates). The last column is a numeric value which is unique to every position.
As an exercise for myself I try to feed this data into the model example from Pytorch, but I keep getting repeating accuracies. In my problem there are two classes of graphs that are differentiated based on the density of points (particularly at the beginning and end), I’ve added plots below as an example. I’ve taken relative distances between nodes to be an edge attribute where each node (excluding the first and last point) has only two undirected connections and the last column in my data as a node feature. Any idea where I may be going wrong is much appreciated!
Below is the slightly modified code provided by the Pytorch example
class GCN(torch.nn.Module):
def __init__(self, hidden_channels,num_classes,num_node_features):
super(GCN, self).__init__()
#torch.manual_seed(12345)
self.conv1 = GCNConv(num_node_features, hidden_channels)
self.conv2 = GCNConv(hidden_channels, hidden_channels)
self.conv3 = GCNConv(hidden_channels, hidden_channels)
self.lin = Linear(hidden_channels,num_classes)#, dataset.num_classes)
def forward(self, x, edge_index, batch,edge_attr=None):
# 1. Obtain node embeddings
x = self.conv1(x, edge_index, edge_attr)
x = x.relu()
x = self.conv2(x, edge_index, edge_attr)
x = x.relu()
x = self.conv3(x, edge_index, edge_attr)
# 2. Readout layer
x = global_mean_pool(x, batch) # [batch_size, hidden_channels]
# 3. Apply a final classifier
x = F.dropout(x, p=0.25, training=self.training)
x = self.lin(x)
return x
def train():
model.train()
for data in train_loader: # Iterate in batches over the training dataset.
#print(data)
out = model(data.x, data.edge_index, data.batch, data.edge_attr) # Perform a single forward pass.
#print(data.y)
loss = criterion(out, data.y) # Compute the loss.
loss.backward() # Derive gradients.
optimizer.step() # Update parameters based on gradients.
optimizer.zero_grad() # Clear gradients.
def test(loader):
model.eval()
correct = 0
for data in loader: # Iterate in batches over the training/test dataset.
out = model(data.x, data.edge_index, data.batch, data.edge_attr)
#for name, param in model.named_parameters():
# if param.requires_grad:
# print(name, param.data)
pred = out.argmax(dim=1) # Use the class with highest probability.
correct += int((pred == data.y).sum()) # Check against ground-truth labels.
return correct / len(loader.dataset) # Derive ratio of correct predictions.
train_loader data:
Step 1:
=======
Number of graphs in the current batch: 64
DataBatch(x=[21826, 1], edge_index=[2, 43524], edge_attr=[43524, 1], y=[64], batch=[21826], ptr=[65])
Step 2:
=======
Number of graphs in the current batch: 64
DataBatch(x=[21970, 1], edge_index=[2, 43812], edge_attr=[43812, 1], y=[64], batch=[21970], ptr=[65])
Step 3:
=======
Number of graphs in the current batch: 64
DataBatch(x=[22798, 1], edge_index=[2, 45468], edge_attr=[45468, 1], y=[64], batch=[22798], ptr=[65])
Step 4:
=======
Number of graphs in the current batch: 64
DataBatch(x=[22130, 1], edge_index=[2, 44132], edge_attr=[44132, 1], y=[64], batch=[22130], ptr=[65])
Step 5:
=======
Number of graphs in the current batch: 64
DataBatch(x=[22533, 1], edge_index=[2, 44938], edge_attr=[44938, 1], y=[64], batch=[22533], ptr=[65])
Step 6:
=======
Number of graphs in the current batch: 64
DataBatch(x=[22167, 1], edge_index=[2, 44206], edge_attr=[44206, 1], y=[64], batch=[22167], ptr=[65])
Step 7:
=======
Number of graphs in the current batch: 64
DataBatch(x=[22017, 1], edge_index=[2, 43906], edge_attr=[43906, 1], y=[64], batch=[22017], ptr=[65])
Step 8:
=======
Number of graphs in the current batch: 64
DataBatch(x=[21578, 1], edge_index=[2, 43028], edge_attr=[43028, 1], y=[64], batch=[21578], ptr=[65])
Step 9:
=======
Number of graphs in the current batch: 64
DataBatch(x=[21597, 1], edge_index=[2, 43066], edge_attr=[43066, 1], y=[64], batch=[21597], ptr=[65])
Step 10:
=======
Number of graphs in the current batch: 64
DataBatch(x=[20545, 1], edge_index=[2, 40962], edge_attr=[40962, 1], y=[64], batch=[20545], ptr=[65])
Step 11:
=======
Number of graphs in the current batch: 64
DataBatch(x=[21283, 1], edge_index=[2, 42438], edge_attr=[42438, 1], y=[64], batch=[21283], ptr=[65])
Step 12:
=======
Number of graphs in the current batch: 64
DataBatch(x=[21038, 1], edge_index=[2, 41948], edge_attr=[41948, 1], y=[64], batch=[21038], ptr=[65])
Step 13:
=======
Number of graphs in the current batch: 64
DataBatch(x=[22283, 1], edge_index=[2, 44438], edge_attr=[44438, 1], y=[64], batch=[22283], ptr=[65])
Step 14:
=======
Number of graphs in the current batch: 64
DataBatch(x=[21734, 1], edge_index=[2, 43340], edge_attr=[43340, 1], y=[64], batch=[21734], ptr=[65])
Step 15:
=======
Number of graphs in the current batch: 64
DataBatch(x=[22095, 1], edge_index=[2, 44062], edge_attr=[44062, 1], y=[64], batch=[22095], ptr=[65])
Step 16:
=======
Number of graphs in the current batch: 64
DataBatch(x=[21877, 1], edge_index=[2, 43626], edge_attr=[43626, 1], y=[64], batch=[21877], ptr=[65])
Step 17:
=======
Number of graphs in the current batch: 64
DataBatch(x=[21465, 1], edge_index=[2, 42802], edge_attr=[42802, 1], y=[64], batch=[21465], ptr=[65])
Step 18:
=======
Number of graphs in the current batch: 64
DataBatch(x=[21314, 1], edge_index=[2, 42500], edge_attr=[42500, 1], y=[64], batch=[21314], ptr=[65])
Step 19:
=======
Number of graphs in the current batch: 64
DataBatch(x=[22256, 1], edge_index=[2, 44384], edge_attr=[44384, 1], y=[64], batch=[22256], ptr=[65])
Step 20:
=======
Number of graphs in the current batch: 25
DataBatch(x=[8761, 1], edge_index=[2, 17472], edge_attr=[17472, 1], y=[25], batch=[8761], ptr=[26])
model = GCN(hidden_channels=64, num_classes=2, num_node_features = 1)
optimizer = torch.optim.Adam(model.parameters(), lr=0.01)
criterion = torch.nn.CrossEntropyLoss()
for epoch in range(1, 171):
train()
train_acc = test(train_loader)
test_acc = test(test_loader)
print(f'Epoch: {epoch:03d}, Train Acc: {train_acc:.4f}, Test Acc: {test_acc:.4f}')
Example output:
Epoch: 001, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 002, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 003, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 004, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 005, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 006, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 007, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 008, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 009, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 010, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 011, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 012, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 013, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 014, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 015, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 016, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 017, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 018, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 019, Train Acc: 0.5947, Test Acc: 0.5942
Epoch: 020, Train Acc: 0.5947, Test Acc: 0.5942