Hi everyone! I am working with somewhat complicated RNN architectures that receive inputs from multiple sources in such a way that requires me to process each RNN layer separately and in a sequential fashion. Because of this, I am utilizing the RNNCell block. However, I’ve noticed that when training these networks, the memory utilization far exceeds those of RNNs with the same architectures. As a simple experiment, I devised a 1-layer RNN using the classic RNN() and compared it to an RNNCell(). This is what my sample code looks like:
For RNN -
class RNN(nn.Module):
def __init__(self, input_size, hidden_size, num_layers, num_classes):
super(RNN, self).__init__()
self.hidden_size = hidden_size
self.num_layers = num_layers
self.rnn = nn.RNN(input_size, hidden_size, num_layers, nonlinearity = 'relu', batch_first = True)
self.fc = nn.Linear(hidden_size, num_classes)
def forward(self, x):
h0 = torch.zeros(self.num_layers, x.size(0), self.hidden_size).to(device)
print(h0.shape)
output, hidden = self.rnn(x, h0) # output shape (batch_size, seq_length, hidden_size)RNN)
output = output[:, -1, :]
output = self.fc(output)
return output
For the analogous RNNCell -
## make the model - a recurrent network with one hidden layer, and a fully connected output layer
class RNN(nn.Module):
def __init__(self, input_size, hidden_size, num_layers, num_classes):
super(RNN, self).__init__()
self.hidden_size = hidden_size
self.num_layers = num_layers
self.rnn = nn.RNNCell(input_size, hidden_size, num_layers, nonlinearity = 'relu')
self.fc = nn.Linear(hidden_size, num_classes)
def forward(self, x):
h = None
for t in range(x.shape[1]):
h = self.rnn(x[:,t,:], h)
output = self.fc(h)
return output
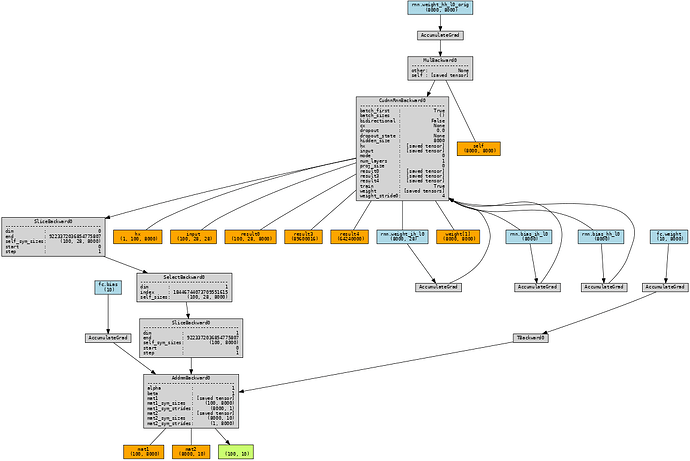
I’ve determined that during validation, the memory usage is quite similar, yet during training the RNN utilizes 3.43GB while the RNNCell uses 8.97GB. I’ve also taken a look at the computational graph using torchviz to determine that the RNN generates one that looks like the following:
while the
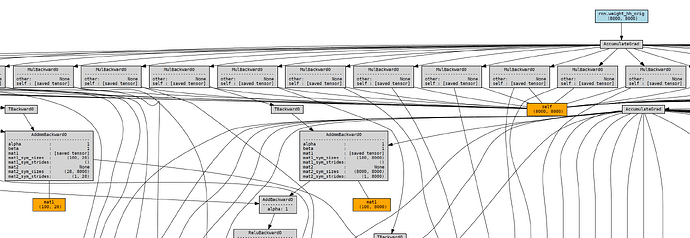
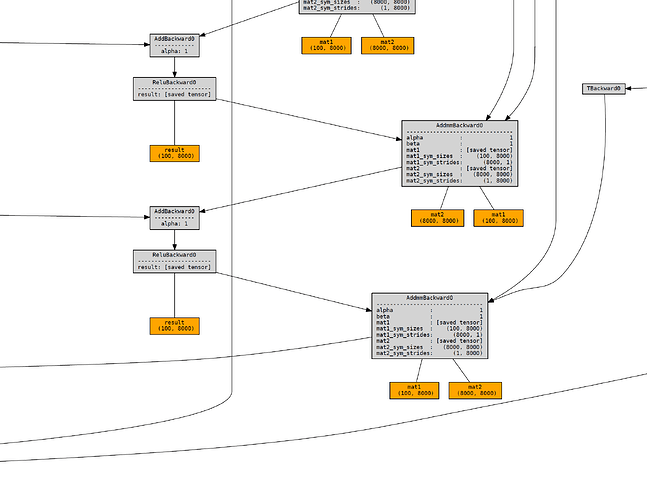
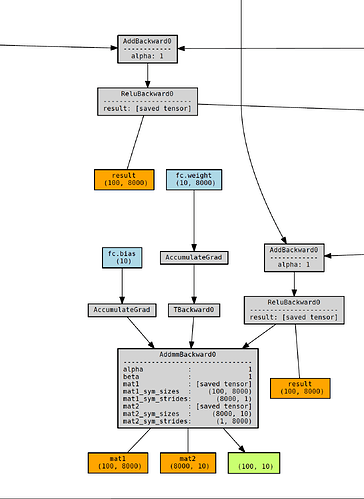
RNNCell generates a much more complicated one, which scales with the sequence length:I know that the RNN() class itself is built on RNNCell(), so I assume some kind of optimization is being done behind the scenes to avoid massive memory usage. I’m aware that PyTorch allows for gradient checkpointing and am wondering if this might be what’s happening within the actual RNN code.
So I guess my questions are:
- How does
RNNmanage to be memory efficient? I don’t believe it scales with sequence length, but I haven’t tested this extensively myself. - How can I make my code more efficient/as efficient as what is possible with
RNN? At the current moment, I am limited on how large my networks can be, even when running them on A100s.
Thanks in advance for any and all feedback!