When dealing with limited Time Series data it can be helpful to apply a sliding window to augment the dataset. However, as I quickly discovered, this can cause data to leak between your training, validation and test sets - especially if you use PyTorch’s built-in random_split(). I expected this to be a common problem, but couldn’t find any discussion on the PyTorch forum nor any ready-to-go methods for preventing overlaps (e.g. in Sklearn). Below I show my attempt at solving this problem, but I am wondering if:
A. There is a standard solution / built-in method for this that I have missed
B. There is a better way to do it
My 1st solution was to do a 80-20 split with contiguous data, to prevent overlap I check the last sample in the 1st split to see if its forecasting horizon overlaps the forecasting horizon of the first sample in the 2nd split. If it does, then I cut those labels in the 2nd split (i.e. we have a small gap between the 2 splits). This also makes cross-validation reasonably easy, since you can just shift the 20% along while taking care of the overlaps.
def split_by_prop(indices:np.array, prop:float, history:int=10, horizon:int=4, fold:int=0):
size = int(len(indices)*prop)
size = size if prop < 0.5 else len(indices) - size
inner_fold = True if fold > 0 else False
size = size + (horizon - 1) if inner_fold else size
offset = indices.min()
splitA = np.arange(offset + fold*size + inner_fold*(horizon - 1),
min(offset + (fold+1)*size - horizon - history + 1, indices.max()+1))
splitB = np.concatenate([np.arange(offset, offset+fold*size), np.arange(offset + (fold+1)*size, indices.max())])
splits = [splitB, splitA]
# Verify that there are no overlaps
for split_no, split in enumerate(splits):
# Get Discontinuity in Split (if there is one)
mask = np.diff(split, append=1) > 1
jump_idx = np.argmax(mask) if sum(mask) >= 1 else None
if jump_idx:
jump_sample = np.arange(split[jump_idx], split[jump_idx] + history)
jump_output = np.arange(split[jump_idx] + history, split[jump_idx] + history + horizon)
# Examine Samples at Boundaries
first_samp = np.arange(split.min(), split.min() + history)
first_output = np.arange(split.min() + history, split.min() + history + horizon)
last_samp = np.arange(split.max(), split.max() + history)
last_output = np.arange(split.max() + history, split.max() + history + horizon)
n_lost_samples = len(indices)-sum([len(split) for split in splits])
return splits[0], splits[1]
However, if your data is sparse, then doing a 80-20 split like this might mean that you have no labels of 1 class in your validation set at all! Alternatively, if your data has strong seasonal patterns, the validation set might not at all be representative of your problem anymore.
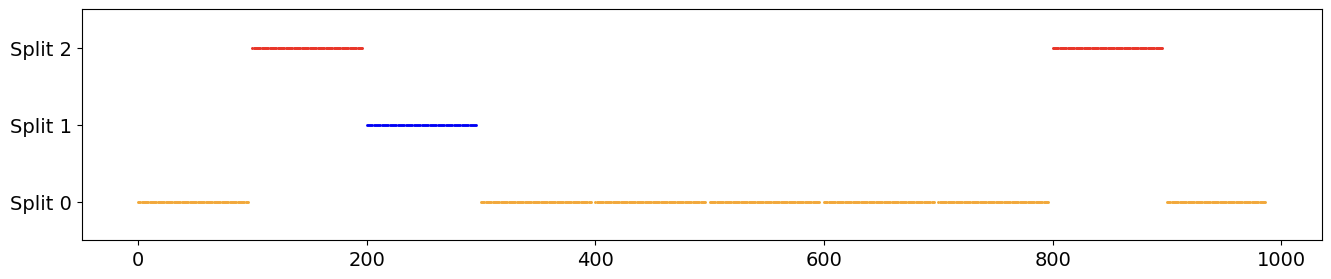
To resolve this I tried to make a “chunking” method, where the timestep ‘indices’ from the original data are split into N chunks without any overlaps. The training, validation and test sets can then be created by choosing a proportion of those chunks:
def split_by_chunks(indices, chunks, train_prop=0.8, val_prop=0.2, internal_seed=0,
fold:int=0, shuffle:bool=False, history:int=10, horizon:int=4):
raw_chunk_size = len(indices) // chunks
offset = history + horizon
chunk_size = raw_chunk_size - horizon + 1
chunk_order = np.arange(chunks)
max_timestamp = len(indices)
if shuffle: # Fixed seed to prevent data leakage
rng = np.random.default_rng(internal_seed)
rng.shuffle(chunk_order)
# Assign chunks to each Split
main_size = int(train_prop*chunks)
val_size = int(val_prop*main_size)
main_chunk_ids = chunk_order[0:main_size]
test_chunk_ids = chunk_order[~np.isin(chunk_order, main_chunk_ids)]
val_chunk_ids = main_chunk_ids[fold*val_size:(fold+1)*val_size]
# Generate and distribute Chunks to each split
train_chunks, test_chunks, val_chunks = [], [], []
for chunk_no in chunk_order:
i = chunk_no * raw_chunk_size
if max_timestamp - offset + 1 <= i:
continue
else:
chunk = np.arange(i, min(i + chunk_size, max_timestamp - offset + 1))
if chunk_no in val_chunk_ids:
val_chunks.append(chunk)
elif chunk_no in test_chunk_ids:
test_chunks.append(chunk)
else:
train_chunks.append(chunk)
return train_chunks, val_chunks, test_chunks
Note: I have removed logging / plotting code to improve readability.
Related Discussions:
Is there a data.Datasets way to use a sliding window over time series data?