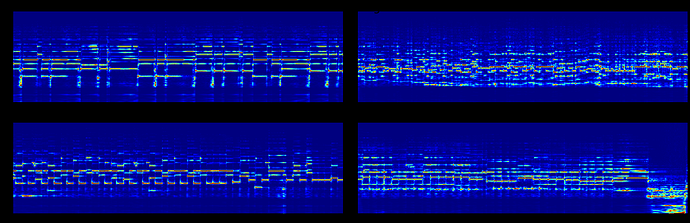
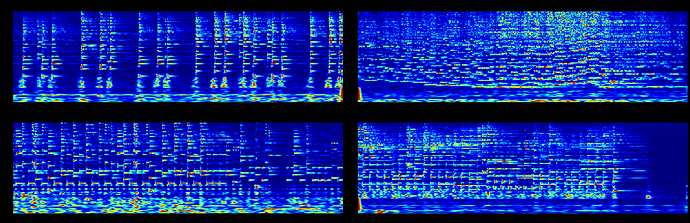
I have few spectrograms which look like this:
x = torch.load('input')
fig, axs = plt.subplots(2, 2, figsize=(24,8))
axs = axs.flat
for idx, i in enumerate(x.cpu().detach().numpy()):
i = scaler.fit_transform(i)
axs[idx].imshow(i, origin='lower', cmap='jet')
axs[idx].axis('off')
fig.tight_layout()
fig.suptitle('Correct image', size=30)
However, my neural network requires some axes swapping along the last two dimension. I realized that swapping the images distort the images,
x = x.transpose(-1,-2)
fig, axs = plt.subplots(2, 2, figsize=(24,8))
axs = axs.flat
for idx, i in enumerate(x.cpu().detach().numpy()):
i = scaler.fit_transform(i)
axs[idx].imshow(i.swapaxes(0,1), origin='lower', cmap='jet')
axs[idx].axis('off')
fig.tight_layout()
fig.suptitle('Wrong image', size=30)
What am I doing wrong here?
You can download my images below
https://sutdapac-my.sharepoint.com/:u:/g/personal/kinwai_cheuk_mymail_sutd_edu_sg/Eb711s15rkFDhcTTUh4vRXkB55rZ3WCY_OS2f0rDsbK8qg?e=SR94eu
Using your code snippet I get the same output for this dummy example:
# Create dummy images
x = torch.zeros(4, 100, 100)
x[0, 25, :] = 1.
x[1, 75, :] = 1.
x[2, :, 25] = 1.
x[3, :, 75] = 1.
fig, axs = plt.subplots(2, 2, figsize=(24,8))
axs = axs.flat
for idx, i in enumerate(x.cpu().detach().numpy()):
axs[idx].imshow(i, origin='lower', cmap='jet')
axs[idx].axis('off')
fig.tight_layout()
fig.suptitle('Correct image', size=30)
x = x.transpose(-1,-2)
fig, axs = plt.subplots(2, 2, figsize=(24,8))
axs = axs.flat
for idx, i in enumerate(x.cpu().detach().numpy()):
axs[idx].imshow(i.swapaxes(0,1), origin='lower', cmap='jet')
axs[idx].axis('off')
fig.tight_layout()
fig.suptitle('Wrong image', size=30)
Hi ptrblck, I realized that there is something wrong with the normalization.
But I don’t understand how swapping the axes affect the normalization. (I am normalizing each of the image to range from 0 to 1)
If you are using sklearn.preprocessing.StandardScaler, note that the input is expected to have the shape [n_samples, n_features], while the mean and var have the shape [n_features,]. Passing your original input and the transposed one would thus result in a different scaling.
1 Like