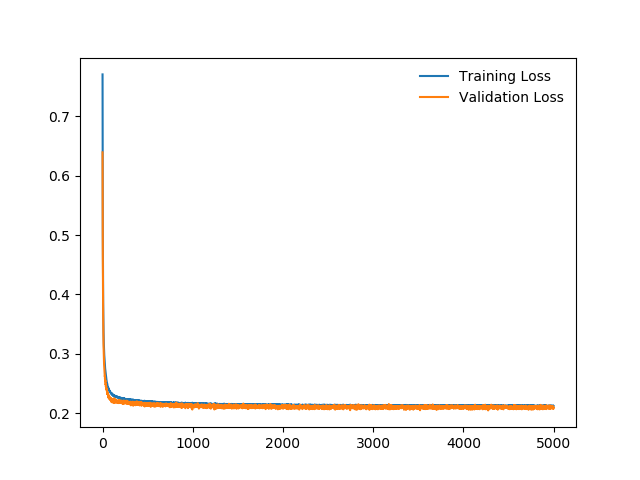
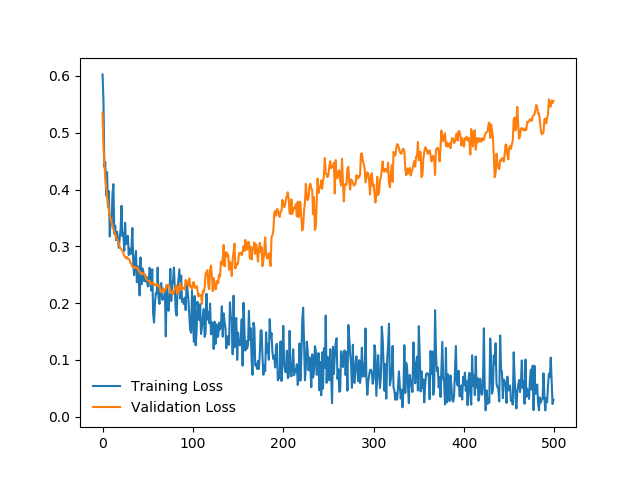
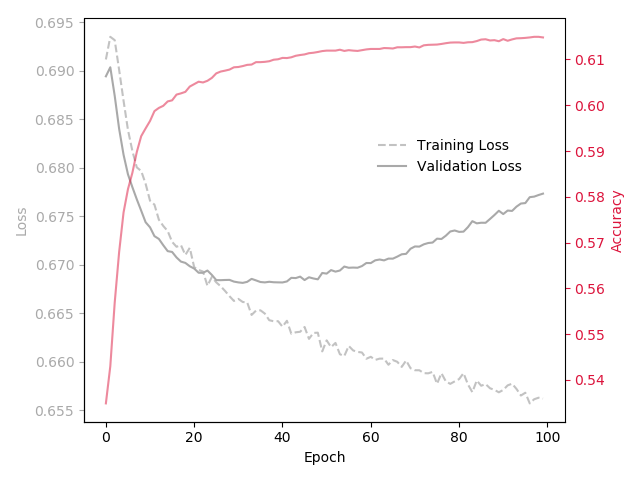
I define this simple network:
class DenseModel(nn.Module):
def __init__(self, inputSize, hiddenSize, outputSize, numLayers, p):
super().__init__()
self.i2h = nn.Linear(inputSize, hiddenSize)
self.relu = nn.ReLU()
self.h2h = nn.Linear(hiddenSize, hiddenSize)
self.h2o = nn.Linear(hiddenSize, outputSize)
self.softmax = nn.Softmax(dim=1) # Normalize output to probs.
self.dropout = nn.Dropout(p)
def forward(self, x):
out = self.i2h(x)
out = self.relu(out)
out = self.dropout(out)
layer = 1
while layer <= numLayers:
out = self.h2h(out)
out = self.relu(out)
out = self.dropout(out)
layer+=1
out = self.h2o(out)
out = self.softmax(out)
return out
Output is two-classes, loss is nn.CrossEntropyLoss, optimizing using sgd like so:
for epoch in range(epochs):
tl = 0
vl = 0
batchCounter = 0
v = 0
for k in trainBatches:
nn1.train()
input_ = Variable(torch.FloatTensor(XTY[k:k+batchSize,:-1]))
target_ = Variable(torch.FloatTensor(XTY[k:k+batchSize,-1:])) # Last position
# Forward
output_ = nn1.forward(input_)
# Backward / Optimize
nn1.zero_grad()
loss = lossFn(output_, torch.max(target_, 1)[0].long())
loss.backward() # Backprop
optimizer.step() # Gradient descent
tl += loss.data.item()
for v in valBatches:
nn1.eval()
input_ = Variable(torch.FloatTensor(XTY[v:v+batchSize,:-1]))
target_ = Variable(torch.FloatTensor(XTY[v:v+batchSize,-1:])) # Last position
# Forward
output_ = nn1.forward(input_)
loss = lossFn(output_, torch.max(target_, 1)[0].long())
vl += loss.data.item()
batchCounter += 1
lossT = np.append(lossT, tl/len(trainBatches)) # Log training loss
lossV = np.append(lossV, vl/len(valBatches)) # Log training loss
My training and validation errors converge, even after 500 epochs:
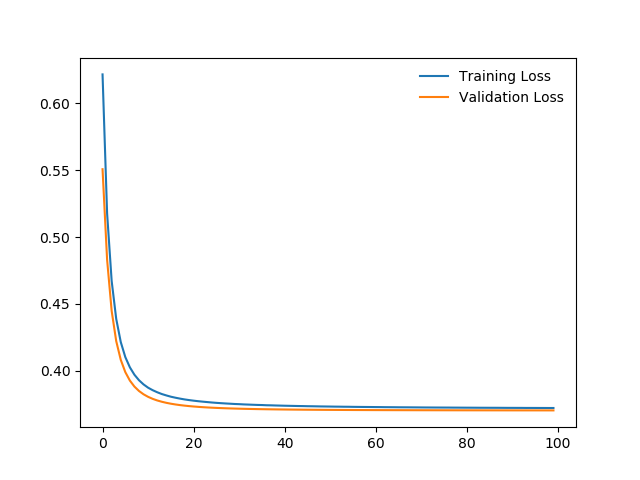
I can’t anything helpful, any ideas?
Thanks.