Hello this is my first post ever so sorry in advance if I don’t meet the standards for posting here.
Context :
I have a problem with training a RNN I try to implement from a paper.
This RNN is called a state-space RNN with 2 equations :
- A state evolution equation which corresponds to the evolution of the hidden state in the network – so we try to learn the evolution of the state. So my assumption is we need not to detach the sate inside the mini batch but we need to initialize it at each new batch. Also we need to retain the graph.
- An output equation : using the current state and input predict the output at the current time step
Below the corresponding equations where x is the hidden state and y the output
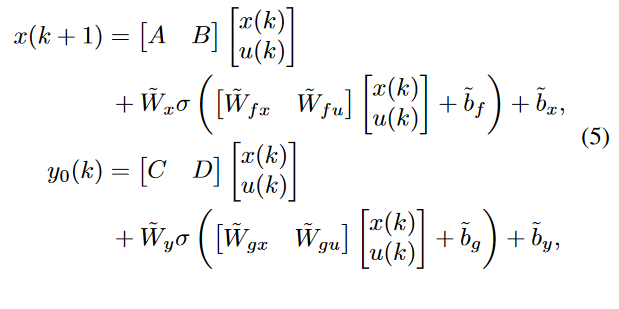
My problem is : when I try to feed to the network each time step k for k from 0 to batch_size. The backpropagation works for the first batch but not for the second…
Giving the classic error : one of the variables needed for gradient computation has been modified by an inplace operation: [torch.FloatTensor [15, 1]], which is output 0 of AsStridedBackward0, is at version 2; expected version 1 instead.
I spent time looking for solutions to this error but : detaching the state does not seem relevant, I do not think there is an inplace operation I can correct in my code.
Please find below the trace given with torch.autograd.set_detect_anomaly(True) set, and my code. I do not understand why the ‘NL2’ layer cannot compute gradient specifically…
Epoch n° 0 started
0
1
C:\Users\Alexandre\anaconda3\envs\grLipNSSM\lib\site-packages\torch\autograd\__init__.py:154: UserWarning: Error detected in AddmmBackward0. Traceback of forward call that caused the error:
File "C:\Users\Alexandre\anaconda3\envs\grLipNSSM\lib\runpy.py", line 194, in _run_module_as_main
return _run_code(code, main_globals, None,
File "C:\Users\Alexandre\anaconda3\envs\grLipNSSM\lib\runpy.py", line 87, in _run_code
exec(code, run_globals)
File "c:\Users\Alexandre\.vscode\extensions\ms-python.python-2021.12.1559732655\pythonFiles\lib\python\debugpy\__main__.py", line 45, in <module>
cli.main()
File "c:\Users\Alexandre\.vscode\extensions\ms-python.python-2021.12.1559732655\pythonFiles\lib\python\debugpy/..\debugpy\server\cli.py", line 444, in main
run()
File "c:\Users\Alexandre\.vscode\extensions\ms-python.python-2021.12.1559732655\pythonFiles\lib\python\debugpy/..\debugpy\server\cli.py", line 285, in run_file
runpy.run_path(target_as_str, run_name=compat.force_str("__main__"))
File "C:\Users\Alexandre\anaconda3\envs\grLipNSSM\lib\runpy.py", line 265, in run_path
return _run_module_code(code, init_globals, run_name,
File "C:\Users\Alexandre\anaconda3\envs\grLipNSSM\lib\runpy.py", line 97, in _run_module_code
_run_code(code, mod_globals, init_globals,
File "C:\Users\Alexandre\anaconda3\envs\grLipNSSM\lib\runpy.py", line 87, in _run_code
exec(code, run_globals)
File "c:\Users\Alexandre\Desktop\Recherche\Simulation\Lip_NSSM\Tuning_grNSSM.py", line 345, in <module>
main()
File "c:\Users\Alexandre\Desktop\Recherche\Simulation\Lip_NSSM\Tuning_grNSSM.py", line 323, in main
loss_course, accuracy_course = train_network(model, train_loader, test_loader, criterion, EPOCHS, LEARNING_RATE)
File "c:\Users\Alexandre\Desktop\Recherche\Simulation\Lip_NSSM\Tuning_grNSSM.py", line 278, in train_network
epoch_loss = model.train_model(train_loader, initial_state, optimizer, criterion)
File "c:\Users\Alexandre\Desktop\Recherche\Simulation\Lip_NSSM\Tuning_grNSSM.py", line 213, in train_model
output, state = self(data[k], state) # Pass data through the network
File "C:\Users\Alexandre\anaconda3\envs\grLipNSSM\lib\site-packages\torch\nn\modules\module.py", line 1102, in _call_impl
return forward_call(*input, **kwargs)
File "c:\Users\Alexandre\Desktop\Recherche\Simulation\Lip_NSSM\Tuning_grNSSM.py", line 124, in forward
x_2 = self.NL2(x_1)
File "C:\Users\Alexandre\anaconda3\envs\grLipNSSM\lib\site-packages\torch\nn\modules\module.py", line 1102, in _call_impl
return forward_call(*input, **kwargs)
File "C:\Users\Alexandre\anaconda3\envs\grLipNSSM\lib\site-packages\torch\nn\modules\linear.py", line 103, in forward
return F.linear(input, self.weight, self.bias)
File "C:\Users\Alexandre\anaconda3\envs\grLipNSSM\lib\site-packages\torch\nn\functional.py", line 1848, in linear
return torch._C._nn.linear(input, weight, bias)
(Triggered internally at ..\torch\csrc\autograd\python_anomaly_mode.cpp:104.)
Variable._execution_engine.run_backward(
Backend QtAgg is interactive backend. Turning interactive mode on.
Init :
def __init__(self, input_dim = INPUT_DIM, hidden_dim = HIDDEN_DIM, state_dim = STATE_DIM, output_dim = OUTPUT_DIM, init_mode = 'random'):
super(grLipNSSM, self).__init__()
# Number of Input neurons
self.input_dim = input_dim
# Number of hidden dimensions
self.hidden_dim = hidden_dim
# Size of the state space
self.state_dim = state_dim
# Number of output dimensions
self.output_dim = output_dim
# Activation functions
self.actF = torch.nn.Tanh()
# Input layer for the state W_u
self.input_layer = nn.Linear(self.input_dim, self.hidden_dim, bias=False)
# First state layer
self.state_layer = nn.Linear(self.state_dim, self.hidden_dim)
# Nonlinear part for the state
self.NL1 = nn.Linear(self.hidden_dim, self.state_dim)
# Residual block
self.residualA = nn.Linear(self.state_dim, self.state_dim, bias= False)
self.residualB = nn.Linear(self.input_dim, self.state_dim, bias= False)
# Output layers
self.input_layer2 = nn.Linear(self.input_dim, self.hidden_dim, bias= False)
self.state_layer2 = nn.Linear(self.state_dim, self.hidden_dim)
self.NL2 = nn.Linear(self.hidden_dim, self.output_dim)
self.residualC = nn.Linear(self.state_dim, self.output_dim, bias= False)
self.residualD = nn.Linear(self.input_dim, self.output_dim, bias= False)
Forward pass
def forward(self, u, state = torch.zeros(1, STATE_DIM)):
# Forward pass -- prediction of the output at time k : y_k
x_0 = self.input_layer2(u)
# Skip connections layers
sc_A = self.residualA(state)
sc_B = self.residualB(u)
sc_C = self.residualC(state)
sc_D = self.residualD(u)
# Hidden layers in the non linear part
h1 = self.state_layer(state)
h2 = self.state_layer2(state)
x_1 = self.actF((x_0 + h2)) #Add another activation function
x_2 = self.NL2(x_1)
y_k = x_2 + sc_C + sc_D
# Updating the state
x_3 = self.input_layer(u)
x_4 = self.actF(x_3+ h1)
x_5 = self.NL1(x_4)
#Adding the linear part (skip connection)
state = x_5 + sc_A + sc_B
return y_k, state
Training function :
def train_model(self, train_loader, initial_state, optimizer, criterion):
"""Train neural network model with Adam optimizer for a single epoch
params:
* model: nn.Sequential instance - NN model to be tested
* train_loader: DataLoader instance - Training data for NN
* initial_state : - Initial state for the eopch
* optimizer: torch.optim instance - Optimizer for NN
* criterion: torch.nn.CrossEntropyLoss instance - Loss function
* state : stat
modifies:
weights of neural network model instance
"""
self.train() # Set model to training mode
sim_loss = 0
epoch_sim_loss = 0
for idx, (data, target) in enumerate(train_loader):
data = data.view(BATCH_SIZE, -1)
state = self.init_hidden()
print(idx)
optimizer.zero_grad() # Zero gradient buffers
for k in range(len(data)): # Iterates through time
output, state = self(data[k], state) # Pass data through the network
sim_loss += criterion(output, target[k]) # Calculate loss
sim_loss.backward(retain_graph=True) # Backpropagate
optimizer.step() # Update weights
epoch_sim_loss += sim_loss.item()
return epoch_sim_loss
The error is occurring when we are doing the backprop step for the 2nd minibatch
Thank you for your help.