I’ve understood the process of labeling for semantic segmentation for 2D images. I was able to create label or target tensors using a colour coded method provided for the dataset. The colour codes provided were:
("Animal", np.array([64, 128, 64], dtype=np.uint8)),
("Archway", np.array([192, 0, 128], dtype=np.uint8)),
("Bicyclist", np.array([0, 128, 192], dtype=np.uint8)),
("Bridge", np.array([0, 128, 64], dtype=np.uint8)),
("Building", np.array([128, 0, 0], dtype=np.uint8)),
("Car", np.array([64, 0, 128], dtype=np.uint8)),
("CartLuggagePram", np.array([64, 0, 192], dtype=np.uint8)),
("Child", np.array([192, 128, 64], dtype=np.uint8)),
To create a label for my network I would simply locate these RGB colours in the images and compare each “object” with its respective colour code and then give it a value in a grayscale image. So each label would be a single channel tensor. And the output of my system would have channels equal to the number of classes in the dataset (for CamVid this was 32).
No I am trying to do the same in 3D and am struggling to understand what to do. The problem is a binary one as I have to detect a single item in the input so basically I should have two output channels: Background and foreground. I have a 3D single channel input, the input having the shape: [BS, Channel, Z, X, Y].
For this input, I have a tensor which indicates the locations of my object of interest in the input. A 3D Gaussian is built around each point to be measured.
I hope I have explained my situation clearly.
My confusion is how to create a label tensor in 3D for segmenting my points of interest as I have done for the CamVid dataset.
Below is a 2D representation of a target that has 3D Gaussians drawn on the points of interest, the tensor is of the size [16, 64, 64]
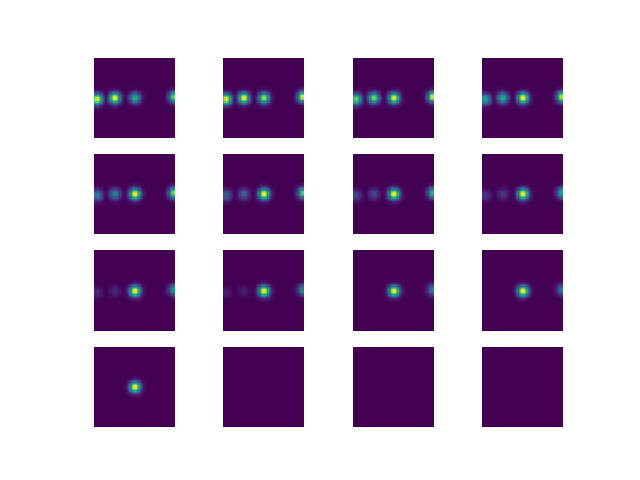
Here is the target file as a pickle dump (just in case): Box
Similar to the CamVid case as I would have 1 channel per class, here I know that I would have 2 channels, one for the background and 1 for the foreground, but how to label these 3D tensors is what I dont understand.

