I’m implementing a neural network to solve ODEs. I want to encode several points at once and get several results of the ODE (one result per corresponding input) at the same run.
Currently I’m running a toy model where I try to get u(x)=sin(x) according to the loss function: du_dx - cos(x).
The network structure is:
network structure
class NetworkClass(nn.Module):
def __init__(self, n_nodes=8):
super().__init__()
self.fc_in = nn.Linear(n_nodes, n_nodes)
self.activation = nn.Tanh()
self.q_layer = nn.Linear(n_nodes, n_nodes)
self.activation_2 = nn.Tanh()
self.fc_out = nn.Linear(n_nodes, n_nodes)
def forward(self, inputs):
out_fc_in = self.fc_in(inputs.unsqueeze(0))
out_activation = self.activation(out_fc_in)
out_q_layer = self.q_layer(out_activation.squeeze(0).squeeze(0))
out_activation_2 = self.activation_2(out_q_layer)
out_fc_out = self.fc_out(out_activation_2).squeeze(0).squeeze(0)
return out_fc_out
The gradients:
gradients
for epoch in range(self.epochs):
u_out_arr = []
d_u_dx_arr = []
loss_single_batch_arr = []
loss_single_batch_no_beta_arr = []
self.opt.zero_grad(set_to_none=False)
collocation_points = torch.reshape(self.collocation_points, [self.n_points//self.n_nodes, self.n_nodes])
u_out = self.model(collocation_points)
u_out_arr.append(u_out) # for plotting and derivative calculation check
d_u_dx = torch.zeros(self.n_points)
for i in range(self.n_batches):
jac_func = jacobian(self.model, collocation_points[i], create_graph=True, strict=True)
d_u_dx[i*self.batch_size:(i+1)*self.batch_size] = torch.diag(jac_func, 0)
d_u_dx_arr.append(d_u_dx)
loss= self.loss_fn(collocation_points, u=torch.flatten(u_out), d_u_dx=d_u_dx,
target_fn=self.target_fn_dict)
loss.backward()
loss_single_batch_arr.append(loss)
self.opt.step()
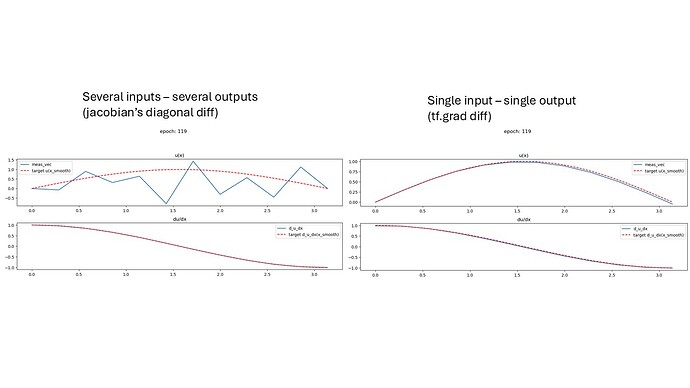
When computing derivatives using jacobian (only the diagonal because I need each output’s derivative with respect to the corresponding input):
- With
in_out_size=1: derivatives correctly correspond to spatial derivatives - With
in_out_size=n_nodes: derivatives don’t match expected spatial derivatives
Question: Why does increasing input/output dimensions affect the derivative computation?
Also, I’ve found that when adding the term of the original_function_loss to the loss_fn in that manner, when beta weights the loss to the original only (beta =1) or the derivative only (beta = 0) or somewhere in between, enables the network to get the accurate results (which is not surprising) by beta as small as 1e-7 (which is very surprising!):
pde_loss_1 = d_u_dx - target_fn["d_u_dx"](torch.flatten(collocation_point))
original_function_loss = u - target_fn["u"](torch.flatten(collocation_point))
mse_loss = torch.mean(self.beta * original_function_loss ** 2 + (1 - self.beta) * pde_loss_1 ** 2)
results for the two cases (without the beta):
Thanks in advance!