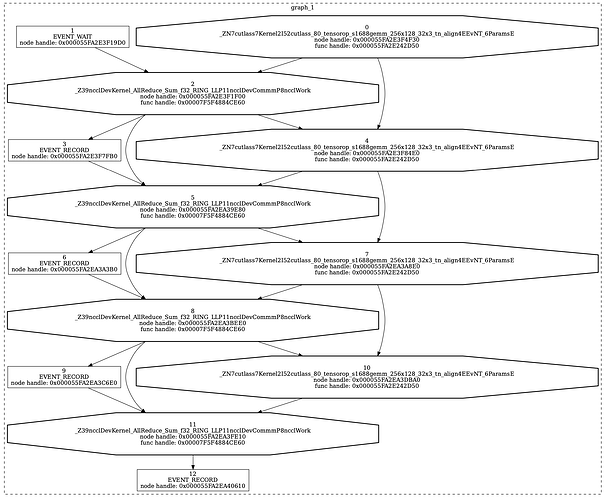
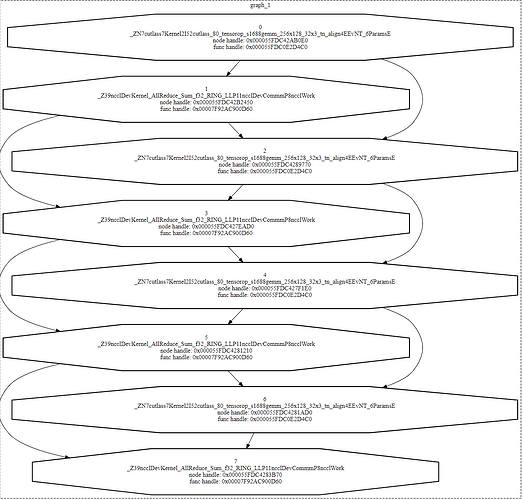
Got a weird one here. I have a small test model that performs a mixture of matmuls and NCCL allreduces to simulate model parallel execution. (Aside: this is the same test code from the open issue at Issues · pytorch/pytorch · GitHub)
import os
import torch
import torch.distributed as dist
class Model(torch.nn.Module):
def __init__(self, dim, nlayers):
super().__init__()
self.layers = torch.nn.ModuleList()
for _ in range(nlayers):
self.layers.append(torch.nn.Linear(dim, dim, bias=False))
@torch.inference_mode()
def forward(self, x):
for layer in self.layers:
x = layer(x)
dist.all_reduce(x)
return x
def main():
os.environ["NCCL_ASYNC_ERROR_HANDLING"] = "0"
dist.init_process_group(backend="nccl")
rank = dist.get_rank()
my_device = f"cuda:{rank}"
torch.cuda.set_device(my_device)
model = Model(2048, 12)
model.cuda()
g = torch.cuda.CUDAGraph()
static_input = torch.empty((1024, 2048), dtype=torch.float32, device=my_device)
s = torch.cuda.Stream()
s.wait_stream(torch.cuda.current_stream())
# Warmup
with torch.cuda.stream(s):
for _ in range(11):
static_output = model(static_input)
torch.cuda.current_stream().wait_stream(s)
with torch.cuda.graph(g):
static_output = model(static_input)
torch.cuda.synchronize()
for _ in range(10):
g.replay()
if __name__ == "__main__":
main()
I run this on two GPUs with torchrun --standalone --nproc_per_node=2 test.py.
What I observe is that the graph replays are consistently longer than I would expect from just summing up matmul and allreduce times. If I generate an nsys profile, the replay region looks like this:
In terms of timing each “layer” is a sequence of:
- matmul: 183us
- GAP: 10us
- allreduce: 44us
It’s that gap I am focused on – we have a production workload (using graphs and nccl) where the gap can be as high as 40us. I can’t figure out what influences the size of the gap, but it’s always present for NCCL allreduces when using CUDA graphs. For vanilla eager execution, you see exactly the profile you’d expect (1-3us between kernels based on whatever).
Looking for pointers, ideas, or anything to try – thanks!