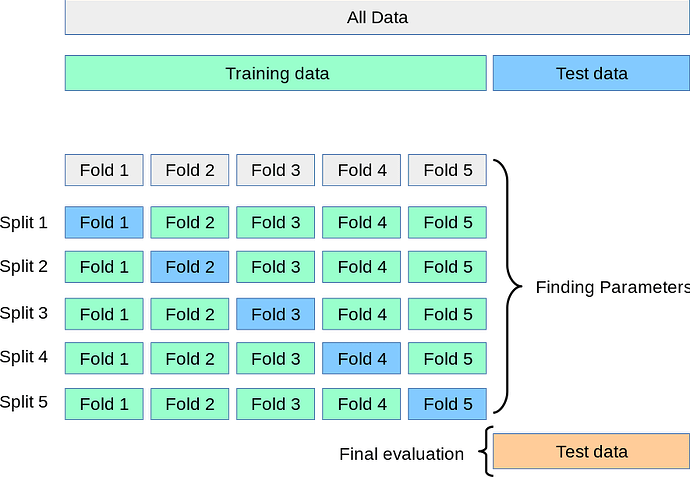
I trained my model using data augmentation, and now I want to use k-fold cross validation to get better result ,so I have to modify the existing script
this is the data function
class VideoDataset(data.Dataset):
def __init__(self,
root_path,
annotation_path,
subset,
spatial_transform=None,
temporal_transform=None,
target_transform=None,
video_loader=None,
video_path_formatter=(lambda root_path, label, video_id:
root_path / label / video_id),
image_name_formatter=lambda x: f'image_{x:05d}.jpg',
target_type='label'):
self.data, self.class_names = self.__make_dataset(
root_path, annotation_path, subset, video_path_formatter)
self.spatial_transform = spatial_transform
self.temporal_transform = temporal_transform
self.target_transform = target_transform
if video_loader is None:
self.loader = VideoLoader(image_name_formatter)
else:
self.loader = video_loader
self.target_type = target_type
def __make_dataset(self, root_path, annotation_path, subset,
video_path_formatter):
with annotation_path.open('r') as f:
data = json.load(f)
video_ids, video_paths, annotations = get_database(
data, subset, root_path, video_path_formatter)
class_to_idx = get_class_labels(data)
idx_to_class = {}
for name, label in class_to_idx.items():
idx_to_class[label] = name
n_videos = len(video_ids)
dataset = []
for i in range(n_videos):
if i % (n_videos // 5) == 0:
print('dataset loading [{}/{}]'.format(i, len(video_ids)))
if 'label' in annotations[i]:
label = annotations[i]['label']
label_id = class_to_idx[label]
else:
label = 'test'
label_id = -1
video_path = video_paths[i]
if not video_path.exists():
continue
segment = annotations[i]['segment']
if segment[1] == 1:
continue
frame_indices = list(range(segment[0], segment[1]))
sample = {
'video': video_path,
'segment': segment,
'frame_indices': frame_indices,
'video_id': video_ids[i],
'label': label_id
}
dataset.append(sample)
return dataset, idx_to_class
def __loading(self, path, frame_indices):
clip = self.loader(path, frame_indices)
if self.spatial_transform is not None:
self.spatial_transform.randomize_parameters()
clip = [self.spatial_transform(img) for img in clip]
clip = torch.stack(clip, 0).permute(1, 0, 2, 3)
return clip
def __getitem__(self, index):
path = self.data[index]['video']
if isinstance(self.target_type, list):
target = [self.data[index][t] for t in self.target_type]
else:
target = self.data[index][self.target_type]
frame_indices = self.data[index]['frame_indices']
if self.temporal_transform is not None:
frame_indices = self.temporal_transform(frame_indices)
clip = self.__loading(path, frame_indices)
if self.target_transform is not None:
target = self.target_transform(target)
return clip, target
def __len__(self):
return len(self.data) `
then in the main function the augmentation were performed
def get_train_utils(opt, model_parameters):
spatial_transform = []
spatial_transform.append(Resize(opt.sample_size))
spatial_transform.append(CenterCrop(opt.sample_size))
normalize = get_normalize_method(opt.mean, opt.std, opt.no_mean_norm,
opt.no_std_norm)
if not opt.no_hflip:
spatial_transform.append(RandomHorizontalFlip())
if not opt.no_vflip:
spatial_transform.append(RandomVerticalFlip())
if opt.colorjitter:
spatial_transform.append(ColorJitter())
spatial_transform.append(ToTensor())
if opt.input_type == 'flow':
spatial_transform.append(PickFirstChannels(n=2))
spatial_transform.append(ScaleValue(opt.value_scale))
spatial_transform.append(normalize)
spatial_transform = Compose(spatial_transform)
assert opt.train_t_crop in ['random','center']
temporal_transform = []
if opt.sample_t_stride > 1:
temporal_transform.append(TemporalSubsampling(opt.sample_t_stride))
if opt.train_t_crop == 'random':
temporal_transform.append(TemporalRandomCrop(opt.sample_duration))
if opt.train_t_crop == 'center':
temporal_transform.append(TemporalCenterCrop(opt.sample_duration))
temporal_transform = TemporalCompose(temporal_transform)
train_data = get_training_data(opt.video_path, opt.annotation_path,
opt.dataset, opt.input_type, opt.file_type,
spatial_transform, temporal_transform)
if opt.distributed:
train_sampler = torch.utils.data.distributed.DistributedSampler(
train_data)
else:
train_sampler = None`Preformatted text`
train_loader = torch.utils.data.DataLoader(train_data,
batch_size=opt.batch_size,
shuffle=(train_sampler is None),
num_workers=opt.n_threads,
pin_memory=True,
sampler=train_sampler,
worker_init_fn=worker_init_fn)
if opt.is_master_node:
train_logger = Logger(opt.result_path / 'train.log',
['epoch', 'loss', 'acc', 'lr'])
train_batch_logger = Logger(
opt.result_path / 'train_batch.log',
['epoch', 'batch', 'iter', 'loss', 'acc', 'lr'])
else:
train_logger = None
train_batch_logger = None
if opt.nesterov:
dampening = 0
else:
dampening = opt.dampening
optimizer = SGD(model_parameters,
lr=opt.learning_rate,
momentum=opt.momentum,
dampening=dampening,
weight_decay=opt.weight_decay,
nesterov=opt.nesterov)
assert opt.lr_scheduler in ['plateau', 'multistep']
assert not (opt.lr_scheduler == 'plateau' and opt.no_val)
if opt.lr_scheduler == 'plateau':
scheduler = lr_scheduler.ReduceLROnPlateau(
optimizer, 'min', patience=opt.plateau_patience)
else:
scheduler = lr_scheduler.MultiStepLR(optimizer,
opt.multistep_milestones)
return (train_loader, train_sampler, train_logger, train_batch_logger,
optimizer, scheduler)
def get_val_utils(opt):
normalize = get_normalize_method(opt.mean, opt.std, opt.no_mean_norm,
opt.no_std_norm)
spatial_transform = [
Resize(opt.sample_size),
CenterCrop(opt.sample_size),
ToTensor()
]
if opt.input_type == 'flow':
spatial_transform.append(PickFirstChannels(n=2))
spatial_transform.extend([ScaleValue(opt.value_scale), normalize])
spatial_transform = Compose(spatial_transform)
temporal_transform = []
if opt.sample_t_stride > 1:
temporal_transform.append(TemporalSubsampling(opt.sample_t_stride))
temporal_transform.append(
TemporalEvenCrop(opt.sample_duration, opt.n_val_samples))
temporal_transform = TemporalCompose(temporal_transform)
val_data, collate_fn = get_validation_data(opt.video_path,
opt.annotation_path, opt.dataset,
opt.input_type, opt.file_type,
spatial_transform,
temporal_transform)
if opt.distributed:
val_sampler = torch.utils.data.distributed.DistributedSampler(
val_data, shuffle=False)
else:
val_sampler = None
val_loader = torch.utils.data.DataLoader(val_data,
batch_size=(opt.batch_size //
opt.n_val_samples),
shuffle=False,
num_workers=opt.n_threads,
pin_memory=True,
sampler=val_sampler,
worker_init_fn=worker_init_fn,
collate_fn=collate_fn)
if opt.is_master_node:
val_logger = Logger(opt.result_path / 'val.log',
['epoch', 'loss', 'acc'])
else:
val_logger = None
return val_loader, val_logger
if I want to use cross validation how can I modify the script and keep the augmentation used can someone guide me on how to do it ?