Hi all,
I’m new to PyTorch and have about 2 months of experience with both PyTorch and MRI data, however, I do have experience in machine learning, primarily with libraries like scikit-learn. I took a deep dive into PyTorch, trying to work with the PI-CAI Challenge dataset (https://pi-cai.grand-challenge.org/) for prostate cancer imaging, which includes T2-weighted images and labels for clinically significant prostate cancer (csPCa).
Summary of My Approach So Far
After some looking around I found that TorchIO (https://torchio.readthedocs.io/) helps organize 3D MRI volumes, apply preprocessing, and create training and testing datasets.
However, I would appreciate feedback on whether this approach is robust and sustainable. Here’s the setup I’ve used (in Google Colab) so far to create a dataset of TorchIO.Subject from PI-CAI images and labels:
!pip install torchio
import os
import torchio as tio
import pandas as pd
# Define paths
base_dir = "/content/drive/MyDrive/picai_public_images_fold0"
csv_path = "/content/drive/MyDrive/marksheet.csv"
# Load the metadata CSV
df = pd.read_csv(csv_path)
df['patient_id'] = df['patient_id'].astype(str)
df.set_index('patient_id', inplace=True)
# Initialize list to store subjects
subjects = []
# List subdirectories in base_dir
for patient_id in os.listdir(base_dir):
if not patient_id.isdigit():
continue
patient_dir = os.path.join(base_dir, patient_id)
if os.path.isdir(patient_dir) and patient_id in df.index:
csPca_label_row = df.loc[patient_id, 'case_csPCa']
csPca_label = 1 if csPca_label_row == 'YES' else 0
# Find the T2W file
t2w_path = next((os.path.join(patient_dir, f) for f in os.listdir(patient_dir) if 't2w.mha' in f), None)
if t2w_path:
subject = tio.Subject(
t2w=tio.ScalarImage(t2w_path),
label=csPca_label,
patient_id=patient_id
)
subjects.append(subject)
else:
print(f"T2W MRI not found for patient {patient_id}")
else:
print(f"Patient {patient_id} is missing from the DataFrame or directory.")
# Checking if subjects loaded correctly
if subjects:
print(f"{len(subjects)} subjects loaded successfully.")
subjects_dataset = tio.SubjectsDataset(subjects)
else:
print("No subjects were created. Please check file paths and keywords.")
Questions & Issues
- Loading Multiple Series: I also used
TorchIOto load DICOM files, but I noticed that TorchIO only loads one series (e.g., T2) if I pass a directory with mixed series (T2, T1, diffusion). However, separating T2, T1, and diffusion DICOMs into different directories works fine. Does this sound like a solid approach, or is there a better way to handle multiple series within a single directory? - Rotated Series: I tried viewing the same series in
Horosand3D Slicer, but the series appears rotated in both. Has anyone encountered this with TorchIO, and is there a solution? - Dataset Splitting: For splitting into train and test sets, I used
sklearn.model_selection.train_test_split, and this appears straightforward. Would you recommend other tools or techniques to ensure balanced splitting with labels? - Unwrapping Tensors: If I need to access each 2D slice in a 3D tensor, what’s the best way to split the tensor along the z-axis? I’d like to explore specific slices within a 3D volume for model input.
- Experience with TorchIO: So far, I find TorchIO promising. Does this setup seem scalable for large MRI datasets like PI-CAI, or am I missing anything critical for 3D data preparation?
I’m not even sure I’m asking the right questions, but I was very relieved to find torchio for this project as my experience is limited.
Here are a few other things I tried:
# Example of accessing the first subject's attributes
first_subject = subjects_dataset[0]
print("Patient ID:", first_subject['patient_id'])
print("Diagnosis (csPCa):", first_subject['label'])
print("T2W Image:", first_subject['t2w'])
Output:
Patient ID: 10648
Diagnosis (csPCa): 0
T2W Image: ScalarImage(shape: (1, 640, 640, 19); spacing: (0.30, 0.30, 3.60); orientation: LPS+; dtype: torch.IntTensor; memory: 29.7 MiB)
Then
first_subject.plot()
Yields
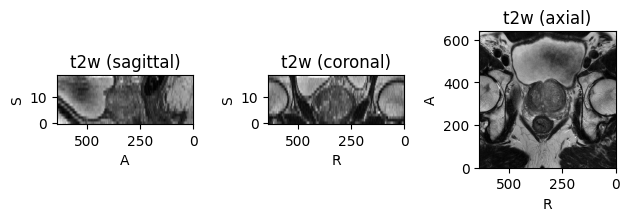
I also tried this:
import matplotlib.pyplot as plt
import torchio as tio
def plot_slices_with_positions(image, spacing, modality="T2W"):
"""Plot each slice in the 3D MRI volume and print its calculated z position."""
# Access the image tensor data, ignoring the channel dimension
img_data = image.data[0] # shape is (640, 640, 19) in this example
# Retrieve spacing for each dimension
spacing_x, spacing_y, spacing_z = spacing
# Determine the number of slices along the depth axis
num_slices = img_data.shape[2]
# Plot each slice with calculated z position
for slice_idx in range(num_slices):
z_position = slice_idx * spacing_z # Calculate the z position based on slice index
plt.imshow(img_data[:, :, slice_idx], cmap='gray')
plt.title(f"{modality} Slice {slice_idx + 1}/{num_slices}\n"
f"X={spacing_x} mm, Y={spacing_y} mm, Z Position={z_position:.2f} mm")
plt.axis('off')
print(f"Slice {slice_idx + 1} - Z Position: {z_position:.2f} mm")
plt.show()
# Example usage on the first subject's T2W image
first_subject = subjects_dataset[0]
t2w_image = first_subject['t2w']
spacing_info = t2w_image.spacing
plot_slices_with_positions(t2w_image, spacing=spacing_info, modality="T2W")
Which yields all slices for example:
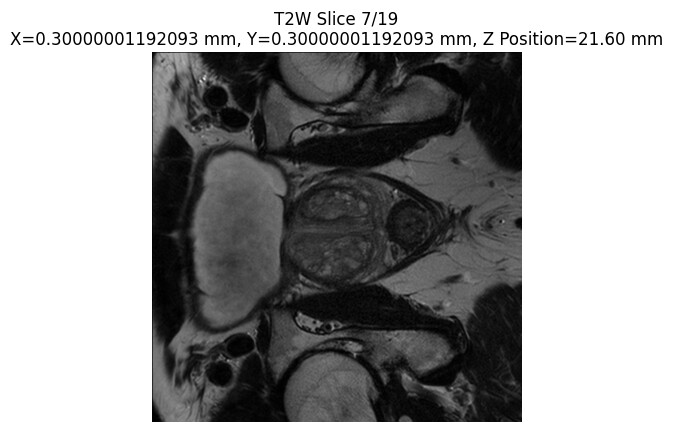
This example is related to what I want to do with extracting 2D slices. Thank you in advance for any insights! I hope it is ok to post this here!