Hi,
I’m doing a medical image segmentation task, and using dice score to validate the performance of my model after every epoch. Everything is going as expected (i.e. training is converging, results look good), except for the fact that my validation set dice score seems to vary lot based on my validation loader batch size.
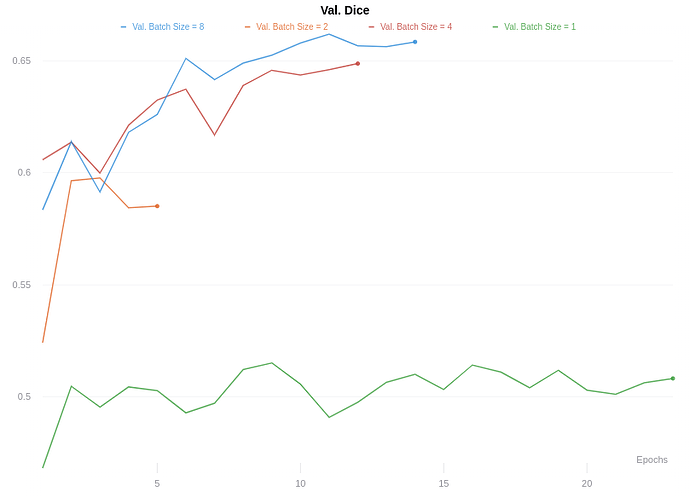
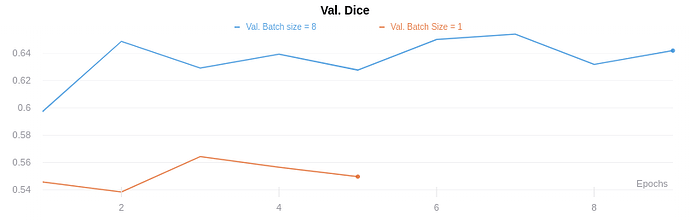
When using a validation batch size of 1, my average dice score is ~0.5, but when I change the validation batch size to 8, it jumps to about 0.65 after 10 epochs. My understanding is the validation accuracy should be independent of the validation set batch sized used.
My training and validation code is as follows (UNet is my U-Net model):
for epoch in range(epochs):
epoch_loss = 0
#Train model
for i_batch, data in enumerate(dataset_loader, 0):
unet.train()
img_batch = data[0].to(device)
true_masks_batch = data[1].to(device)
unet.zero_grad()
mask_pred = unet(img_batch)
loss = DiceDistanceCriterion(mask_pred, true_masks_batch)
epoch_loss += loss.item()
optimizer.zero_grad()
loss.backward()
optimizer.step()
if i_batch % 50 == 0:
train_dice_score = dice_score(mask_pred, true_masks_batch)
print(train_dice_score)
#Validate after each epoch
with torch.no_grad():
val_dice_score = 0
epoch_val_loss = 0
for val_batch, val_sample in enumerate(val_dataset_loader,0):
unet.eval()
val_img = val_sample[0].to(device)
val_mask = val_sample[1].to(device)
pred_val_mask = unet(val_img)
val_loss = DiceDistanceCriterion(pred_val_mask, val_mask)
epoch_val_loss += val_loss.item()
val_dice_score += dice_score(pred_val_mask, val_mask)
if val_batch % 50 == 0:
print(val_dice_score/(val_batch+1))
epoch_loss = epoch_loss/(i_batch + 1)
epoch_val_loss = epoch_val_loss/(val_batch + 1)
val_dice_score = val_dice_score/(val_batch + 1)
print('Epoch: {}/{} --- Val. Loss: {}'.format(epoch, epochs, epoch_val_loss))
scheduler.step(epoch_loss)
And dice score is defined as:
def dice_score(input, target):
eps = 1e-8
iflat = input.view(-1)
tflat = target.view(-1)
intersection = (iflat * tflat).sum()
return ((2. * intersection + eps) /
(iflat.sum() + tflat.sum() + eps))
But my validation dice score looks like this, as you can see it changes largely based on batch size.