Hi ,
train_df.shape
(7111, 8)
I have following predictor variable
deg_C relative_humidity absolute_humidity sensor_1 sensor_2 sensor_3 sensor_4 sensor_5
0 13.1 46.0 0.7578 1387.2 1087.8 1056.0 1742.8 1293.4
1 13.2 45.3 0.7255 1279.1 888.2 1197.5 1449.9 1010.9
and here target variables
target_carbon_monoxide target_benzene target_nitrogen_oxides
0 2.5 12.0 167.7
1 2.1 9.9 98.9
My workaround is
Scaling Both Predictor and targets
X_train, X_val, y_train, y_val = train_test_split(train_df, train_y, test_size=0.2, random_state=0)
scalerX = StandardScaler().fit(X_train)
scalery = StandardScaler().fit(y_train)
X_train = scalerX.transform(X_train)
y_train = scalery.transform(y_train)
X_val = scalerX.transform(X_val)
y_val = scalery.transform(y_val)
class ClassifierDataset(Dataset):
def __init__(self, X_data, y_data):
self.X_data = X_data
self.y_data = y_data
def __getitem__(self, index):
return self.X_data[index], self.y_data[index]
def __len__ (self):
return len(self.X_data)
NUM_FEATURES = 8
BATCHSIZE = 500
train_dataset = ClassifierDataset(torch.from_numpy(X_train), torch.from_numpy(y_train))
train_loader = DataLoader(dataset=train_dataset,batch_size=BATCHSIZE)
valid_dataset = ClassifierDataset(torch.from_numpy(X_val), torch.from_numpy(y_val))
valid_loader = DataLoader(dataset=valid_dataset,batch_size=BATCHSIZE)
Model
class multiNet(nn.Module):
def __init__(self, num_feature):
super().__init__()
self.lin0 = nn.Linear(num_feature,5)
self.lin1 = nn.Linear(5, 4)
self.lin2 = nn.Linear(4, 3)
self.bn0 = nn.BatchNorm1d(num_feature)
self.bn1 = nn.BatchNorm1d(5)
self.bn2 = nn.BatchNorm1d(4)
def forward(self, x):
x = self.bn0(x)
x = self.lin0(x)
x = F.relu(x)
x = self.bn1(x)
x = self.lin1(x)
x = F.relu(x)
x = self.lin2(x) # output layer
return x
Optimizer
def get_optimizer(model, lr):
optimizer = torch.optim.Adam(model.parameters(),lr=lr,weight_decay=0.01)
scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(optimizer,patience=10,factor=0.1,threshold=1e1)
return optimizer,scheduler
Weights
def init_weights(m):
if type(m) == nn.Linear:
'''
This is an example i created as instructed by Andrew Ng course
torch.ones([10, 6])*torch.sqrt(torch.Tensor([2.])/torch.Tensor([1.4142]))
'''
m.weight.data = torch.randn(m.weight.size())*torch.sqrt(torch.Tensor([2.])/m.weight.size()[1])
Model Training
seed = 4
torch.manual_seed(seed)
def train_loop(model, epochs, lr):
total = 0
sum_loss = 0
output = 0
criterion = nn.L1Loss()
optim,scheduler = get_optimizer(model, lr = lr)
train_loss_values = []
val_loss_values = []
for epoch in range(epochs):
train_running_loss = 0.0
val_running_loss = 0.0
for data, target in train_loader:
model.train()
output = model(data)
loss = torch.sqrt(criterion(output, target)) # as i need RMSLE
optim.zero_grad()
loss.backward()
optim.step()
train_running_loss =+ loss.item()
if not scheduler.__class__ == torch.optim.lr_scheduler.ReduceLROnPlateau:
scheduler.step()
train_loss_values.append(train_running_loss)
for valid_data, valid_target in valid_loader:
model.eval()
valid_output = model(valid_data)
valid_loss = criterion(valid_output,valid_target)
val_running_loss =+ valid_loss.item()
val_loss_values.append(val_running_loss)
if epoch % 100 ==0:
print(f'epoch : {epoch+1},training loss : {loss} , validation loss : {valid_loss}')
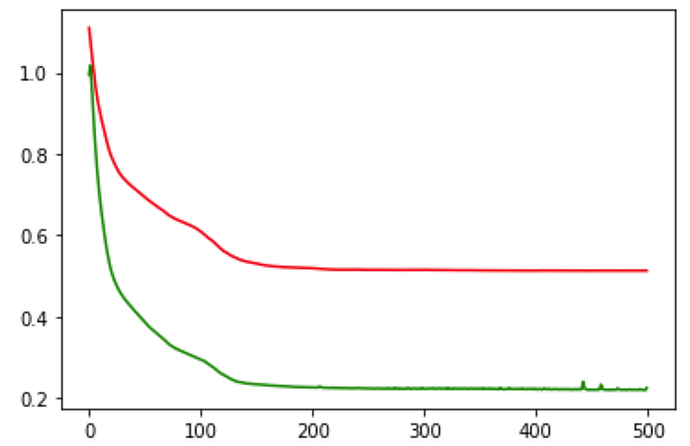
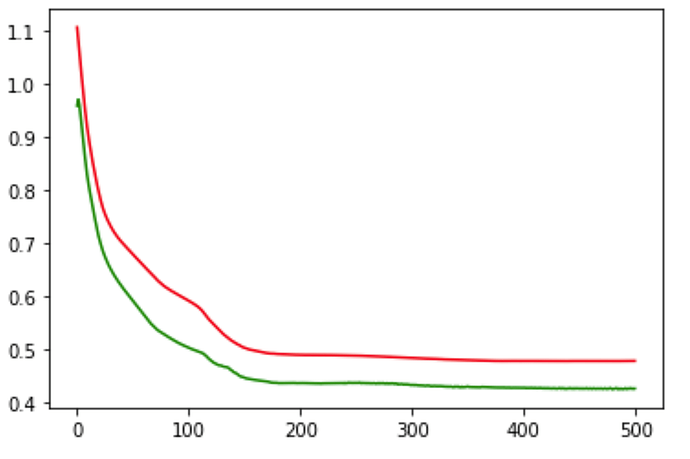
plt.plot(train_loss_values, 'r')
plt.plot(val_loss_values, 'g')
NUM_FEATURES = X_train.shape[1]
multiNetModel = multiNet(NUM_FEATURES)
multiNetModel.apply(init_weights)
train_loop(multiNetModel, epochs=500, lr=0.001)
Is my above baseline approach is correct ? If it is then why my validation loss is way lesser than training loss
epoch : 1 ,training loss : 1.109604 , validation loss : 0.995102
epoch : 101,training loss : 0.608683 , validation loss : 0.294198
epoch : 201,training loss : 0.518566 , validation loss : 0.225262
epoch : 301,training loss : 0.514474 , validation loss : 0.221938
epoch : 401,training loss : 0.512781 , validation loss : 0.220481