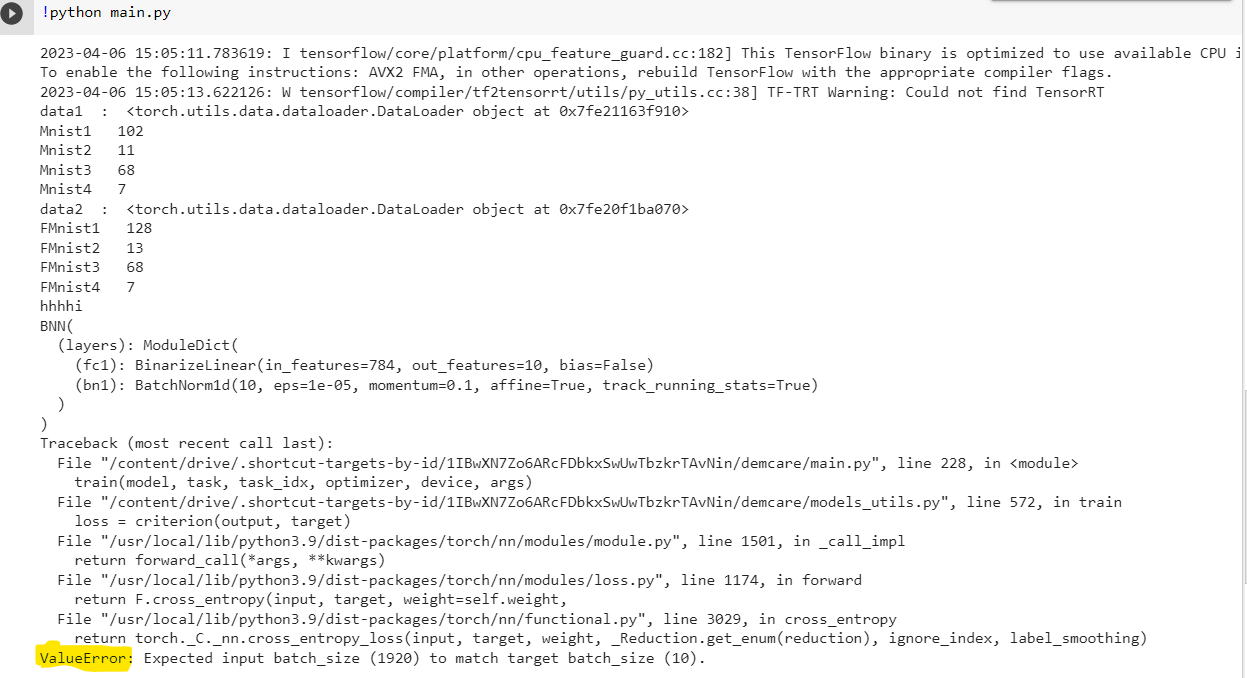
I’m getting the following error
I have tried every of the solution provided on any platform but nothings working. My dataset is demcare(image) . The code Model is pasted below:
class SignActivation(torch.autograd.Function):
@staticmethod
def forward(ctx, i):
result = i.sign()
ctx.save_for_backward(i)
return result
@staticmethod
def backward(ctx, grad_output):
i, = ctx.saved_tensors
grad_i = grad_output.clone()
grad_i[i.abs() > 1.0] = 0
return grad_i
def Binarize(tensor):
return tensor.sign()
class BinarizeLinear(torch.nn.Linear):
def __init__(self, *kargs, **kwargs):
super(BinarizeLinear, self).__init__(*kargs, **kwargs)
def forward(self, input):
if input.size(1) != 784:
input.data=Binarize(input.data)
if not hasattr(self.weight,'org'):
self.weight.org=self.weight.data.clone()
self.weight.data=Binarize(self.weight.org)
out = torch.nn.functional.linear(input, self.weight)
if not self.bias is None:
self.bias.org=self.bias.data.clone()
out += self.bias.view(1, -1).expand_as(out)
return out
class BinarizeConv2d(torch.nn.Conv2d):
def __init__(self, *kargs, **kwargs):
super(BinarizeConv2d, self).__init__(*kargs, **kwargs)
def forward(self, input):
if input.size(1) != 3:
input.data = Binarize(input.data)
if not hasattr(self.weight,'org'):
self.weight.org=self.weight.data.clone()
self.weight.data=Binarize(self.weight.org)
out = torch.nn.functional.conv2d(input, self.weight, None, self.stride,
self.padding, self.dilation, self.groups)
if not self.bias is None:
self.bias.org=self.bias.data.clone()
out += self.bias.view(1, -1, 1, 1).expand_as(out)
return out
class BNN(torch.nn.Module):
def __init__(self, layers_dims, init = "gauss", width = 0.01, norm = 'bn'):
super(BNN, self).__init__()
self.hidden_layers = len(layers_dims)-2
self.layers_dims = layers_dims
self.norm = norm
layer_list = []
for layer in range(self.hidden_layers+1):
layer_list = layer_list + [( ('fc'+str(layer+1) ) , BinarizeLinear(layers_dims[layer], layers_dims[layer+1], bias = False)) ]
if norm == 'bn':
layer_list = layer_list + [( (norm+str(layer+1) ) , torch.nn.BatchNorm1d(layers_dims[layer+1], affine = True, track_running_stats = True)) ]
elif norm == 'in':
layer_list = layer_list + [( (norm+str(layer+1) ) , torch.nn.InstanceNorm1d(layers_dims[layer+1], affine = False, track_running_stats = False)) ]
self.layers = torch.nn.ModuleDict(OrderedDict( layer_list ))
#weight init
for layer in range(self.hidden_layers+1):
if init == "gauss":
torch.nn.init.normal_(self.layers['fc'+str(layer+1)].weight, mean=0, std=width)
if init == "uniform":
torch.nn.init.uniform_(self.layers['fc'+str(layer+1)].weight, a= -width/2, b=width/2)
def forward(self, x):
size = self.layers_dims[0]
x = x.view(-1, size)
for layer in range(self.hidden_layers+1):
x = self.layers['fc'+str(layer+1)](x)
#x = torch.nn.functional.dropout(x, p = 0.5, training = self.training)
if self.norm == 'in': #IN needs channel dim
x.unsqueeze_(1)
x = self.layers[self.norm+str(layer+1)](x)
if self.norm == 'in': #Remove channel dim
x.squeeze_(1)
if layer != self.hidden_layers:
x = SignActivation.apply(x)
return x
def save_bn_states(self):
bn_states = []
if 'bn1' in self.layers.keys():
for l in range(self.hidden_layers+1):
bn = copy.deepcopy(self.layers['bn'+str(l+1)].state_dict())
bn_states.append(bn)
return bn_states
def load_bn_states(self, bn_states):
if 'bn1' in self.layers.keys():
for l in range(self.hidden_layers+1):
self.layers['bn'+str(l+1)].load_state_dict(bn_states[l])
def plot_parameters(model, path, save=True):
fig = plt.figure(figsize=(15, 10))
i = 1
for (n, p) in model.named_parameters():
if (n.find('bias') == -1) and (len(p.size()) != 1): #bias or batchnorm weight -> no plot
fig.add_subplot(2,2,i)
if model.__class__.__name__.find('B') != -1: #BVGG -> plot p.org
if hasattr(p,'org'):
weights = p.org.data.cpu().numpy()
else:
weights = p.data.cpu().numpy()
binet = 100
else:
weights = p.data.cpu().numpy() #TVGG or FVGG plot p
binet = 50
i+=1
plt.title( n.replace('.','_') )
plt.hist( weights.flatten(), binet)
if save:
time = datetime.now().strftime('%H-%M-%S')
fig.savefig(path+'/'+time+'_weight_distribution.png')
plt.close()
class Adam_meta(torch.optim.Optimizer):
def __init__(self, params, lr=1e-3, betas=(0.9, 0.999), meta = {}, eps=1e-8,
weight_decay=0, amsgrad=False):
if not 0.0 <= lr:
raise ValueError("Invalid learning rate: {}".format(lr))
if not 0.0 <= eps:
raise ValueError("Invalid epsilon value: {}".format(eps))
if not 0.0 <= betas[0] < 1.0:
raise ValueError("Invalid beta parameter at index 0: {}".format(betas[0]))
if not 0.0 <= betas[1] < 1.0:
raise ValueError("Invalid beta parameter at index 1: {}".format(betas[1]))
defaults = dict(lr=lr, betas=betas, meta=meta, eps=eps,
weight_decay=weight_decay, amsgrad=amsgrad)
super(Adam_meta, self).__init__(params, defaults)
def __setstate__(self, state):
super(Adam_meta, self).__setstate__(state)
for group in self.param_groups:
group.setdefault('amsgrad', False)
def step(self, closure=None):
loss = None
if closure is not None:
loss = closure()
for group in self.param_groups:
for p in group['params']:
if p.grad is None:
continue
grad = p.grad.data
if grad.is_sparse:
raise RuntimeError('Adam does not support sparse gradients, please consider SparseAdam instead')
amsgrad = group['amsgrad']
state = self.state[p]
if len(state) == 0:
state['step'] = 0
state['exp_avg'] = torch.zeros_like(p.data)
state['exp_avg_sq'] = torch.zeros_like(p.data)
if amsgrad:
# Maintains max of all exp. moving avg. of sq. grad. values
state['max_exp_avg_sq'] = torch.zeros_like(p.data)
exp_avg, exp_avg_sq = state['exp_avg'], state['exp_avg_sq']
if amsgrad:
max_exp_avg_sq = state['max_exp_avg_sq']
beta1, beta2 = group['betas']
state['step'] += 1
if group['weight_decay'] != 0:
grad.add_(group['weight_decay'], p.data)
# Decay the first and second moment running average coefficient
exp_avg.mul_(beta1).add_(1 - beta1, grad)
exp_avg_sq.mul_(beta2).addcmul_(1 - beta2, grad, grad)
if amsgrad:
# Maintains the maximum of all 2nd moment running avg. till now
torch.max(max_exp_avg_sq, exp_avg_sq, out=max_exp_avg_sq)
# Use the max. for normalizing running avg. of gradient
denom = max_exp_avg_sq.sqrt().add_(group['eps'])
else:
denom = exp_avg_sq.sqrt().add_(group['eps'])
bias_correction1 = 1 - beta1 ** state['step']
bias_correction2 = 1 - beta2 ** state['step']
step_size = group['lr'] * math.sqrt(bias_correction2) / bias_correction1
binary_weight_before_update = torch.sign(p.data)
condition_consolidation = (torch.mul(binary_weight_before_update, exp_avg) > 0.0 ) # exp_avg has the same sign as exp_avg/denom
if p.dim()==1: # True if p is bias, false if p is weight
p.data.addcdiv_(-step_size, exp_avg, denom)
else:
decayed_exp_avg = torch.mul(torch.ones_like(p.data)-torch.pow(torch.tanh(group['meta'][p.newname]*torch.abs(p.data)),2), exp_avg)
p.data.addcdiv_(-step_size, torch.where(condition_consolidation, decayed_exp_avg, exp_avg), denom) #assymetric lr for metaplasticity
return loss
class Adam_bk(torch.optim.Optimizer):
def __init__(self, params, lr=1e-3, betas=(0.9, 0.999), n_bk=1, ratios=[0], areas=[1], meta = 0.0, feedback=0.0, eps=1e-8,
weight_decay=0, amsgrad=False, path='.'):
if not 0.0 <= lr:
raise ValueError("Invalid learning rate: {}".format(lr))
if not 0.0 <= eps:
raise ValueError("Invalid epsilon value: {}".format(eps))
if not 0.0 <= betas[0] < 1.0:
raise ValueError("Invalid beta parameter at index 0: {}".format(betas[0]))
if not 0.0 <= betas[1] < 1.0:
raise ValueError("Invalid beta parameter at index 1: {}".format(betas[1]))
defaults = dict(lr=lr, betas=betas, n_bk=n_bk, ratios=ratios, areas=areas, meta=meta, feedback=feedback, eps=eps,
weight_decay=weight_decay, amsgrad=amsgrad, path=path)
super(Adam_bk, self).__init__(params, defaults)
def __setstate__(self, state):
super(Adam_bk, self).__setstate__(state)
for group in self.param_groups:
group.setdefault('amsgrad', False)
def step(self, closure=None):
loss = None
if closure is not None:
loss = closure()
for group in self.param_groups:
n_bk = group['n_bk']
ratios = group['ratios']
areas = group['areas']
meta = group['meta']
feedback = group['feedback']
path = group['path']
for p in group['params']:
if p.grad is None:
continue
grad = p.grad.data
if grad.is_sparse:
raise RuntimeError('Adam does not support sparse gradients, please consider SparseAdam instead')
amsgrad = group['amsgrad']
state = self.state[p]
if len(state) == 0:
state['step'] = 0
state['exp_avg'] = torch.zeros_like(p.data)
state['exp_avg_sq'] = torch.zeros_like(p.data)
# Initializing beakers
for bk_idx in range(n_bk+1):
if bk_idx==n_bk: # create an additional beaker clamped at 0
state['bk'+str(bk_idx)+'_t-1'] = torch.zeros_like(p)
state['bk'+str(bk_idx)+'_t'] = torch.zeros_like(p)
else: # create other beakers at equilibrium
state['bk'+str(bk_idx)+'_t-1'] = torch.empty_like(p).copy_(p)
state['bk'+str(bk_idx)+'_t'] = torch.empty_like(p).copy_(p)
state['bk'+str(bk_idx)+'_lvl'] = []
if amsgrad:
# Maintains max of all exp. moving avg. of sq. grad. values
state['max_exp_avg_sq'] = torch.zeros_like(p.data)
exp_avg, exp_avg_sq = state['exp_avg'], state['exp_avg_sq']
if amsgrad:
max_exp_avg_sq = state['max_exp_avg_sq']
beta1, beta2 = group['betas']
state['step'] += 1
if group['weight_decay'] != 0:
grad.add_(group['weight_decay'], p.data) #p.data
exp_avg.mul_(beta1).add_(1 - beta1, grad)
exp_avg_sq.mul_(beta2).addcmul_(1 - beta2, grad, grad)
if amsgrad:
torch.max(max_exp_avg_sq, exp_avg_sq, out=max_exp_avg_sq)
denom = max_exp_avg_sq.sqrt().add_(group['eps'])
else:
denom = exp_avg_sq.sqrt().add_(group['eps'])
bias_correction1 = 1 - beta1 ** state['step']
bias_correction2 = 1 - beta2 ** state['step']
step_size = group['lr'] * math.sqrt(bias_correction2) / bias_correction1
if p.dim()==1: # True if p is bias, false if p is weight
p.data.addcdiv_(-step_size, exp_avg, denom)
else:
# weight update
p.data.addcdiv_(-step_size, exp_avg, denom)
p.data.add_((ratios[0]/areas[0])*(state['bk1_t-1']-state['bk0_t-1']))
p.data.add_(torch.where( (state['bk'+str(n_bk-1)+'_t-1'] - state['bk0_t-1']) * state['bk'+str(n_bk-1)+'_t-1'].sign() > 0 , feedback*(state['bk'+str(n_bk-1)+'_t-1'] - state['bk0_t-1']),
torch.zeros_like(p.data)))
with torch.no_grad():
for bk_idx in range(1, n_bk):
# diffusion entre les bk dans les deux sens + metaplasticité sur le dernier
if bk_idx==(n_bk-1):
condition = (state['bk'+str(bk_idx-1)+'_t-1'] - state['bk'+str(bk_idx)+'_t-1'])*state['bk'+str(bk_idx)+'_t-1'] < 0
decayed_m = 1 - torch.tanh(meta[p.newname]*state['bk'+str(bk_idx)+'_t-1'])**2
state['bk'+str(bk_idx)+'_t'] = torch.where(condition, state['bk'+str(bk_idx)+'_t-1'] + (ratios[bk_idx-1]/areas[bk_idx])*decayed_m*(state['bk'+str(bk_idx-1)+'_t-1'] - state['bk'+str(bk_idx)+'_t-1']) + (ratios[bk_idx]/areas[bk_idx])*(state['bk'+str(bk_idx+1)+'_t-1'] - state['bk'+str(bk_idx)+'_t-1']),
state['bk'+str(bk_idx)+'_t-1'] + (ratios[bk_idx-1]/areas[bk_idx])*(state['bk'+str(bk_idx-1)+'_t-1'] - state['bk'+str(bk_idx)+'_t-1']) + (ratios[bk_idx]/areas[bk_idx])*(state['bk'+str(bk_idx+1)+'_t-1'] - state['bk'+str(bk_idx)+'_t-1']))
else:
state['bk'+str(bk_idx)+'_t'] = state['bk'+str(bk_idx)+'_t-1'] + (ratios[bk_idx-1]/areas[bk_idx])*(state['bk'+str(bk_idx-1)+'_t-1'] - state['bk'+str(bk_idx)+'_t-1']) + (ratios[bk_idx]/areas[bk_idx])*(state['bk'+str(bk_idx+1)+'_t-1'] - state['bk'+str(bk_idx)+'_t-1'])
fig = plt.figure(figsize=(12,9))
for bk_idx in range(n_bk):
if bk_idx==0:
state['bk'+str(bk_idx)+'_t-1'] = p.data
else:
state['bk'+str(bk_idx)+'_t-1'] = state['bk'+str(bk_idx)+'_t']
if p.size() == torch.empty(4096,4096).size() :
state['bk'+str(bk_idx)+'_lvl'].append(state['bk'+str(bk_idx)+'_t-1'][11, 100].detach().item())
if state['step']%600==0:
plt.plot(state['bk'+str(bk_idx)+'_lvl'])
fig.savefig(path + '/trajectory.png', fmt='png', dpi=300)
plt.close()
if p.dim()!=1 and state['step']%600==0:
fig2 = plt.figure(figsize=(12,9))
for bk_idx in range(n_bk):
plt.hist(state['bk'+str(bk_idx)+'_t-1'].detach().cpu().numpy().flatten(), 100, label='bk'+str(bk_idx), alpha=0.5)
plt.legend()
fig2.savefig(path+'/bk_'+str(bk_idx)+'_'+str(p.size(0))+'-'+str(p.size(1))+'_task'+str((state['step']//48000)%2)+'.png', fmt='png')
torch.save(state, path + '/state_'+str(p.size(0))+'-'+str(p.size(1))+'_task'+str((state['step']//48000)%2)+'.tar')
plt.close()
return loss
def train(model, train_loader, current_task_index, optimizer, device, args,
prev_cons=None, prev_params=None, path_integ=None, criterion = torch.nn.CrossEntropyLoss()):
model.train()
for data, target in train_loader:
if torch.cuda.is_available():
data, target = data.to(device), target.to(device)
optimizer.zero_grad()
output = model(data)
loss = criterion(output, target)
if args.ewc:
ewc_loss = EWC_loss(model, prev_cons, prev_params, current_task_index, device, ewc_lambda=args.ewc_lambda)
total_loss = loss + ewc_loss
elif args.si:
p_prev, p_old = prev_params
si_loss = SI_loss(model, prev_cons, p_prev, args.si_lambda)
total_loss = loss + si_loss
else:
total_loss = loss
total_loss.backward()
for p in list(model.parameters()): # blocking weights with org value greater than a threshold by setting grad to 0
if hasattr(p,'org'):
p.data.copy_(p.org)
optimizer.step()
if args.si:
update_W(model, path_integ, p_old, args)
for p in list(model.parameters()): # updating the org attribute
if hasattr(p,'org'):
p.org.copy_(p.data)
def test(model, test_loader, device, criterion = torch.nn.CrossEntropyLoss(reduction=‘sum’), verbose = False):
model.eval()
test_loss = 0
correct = 0
for data, target in test_loader:
if torch.cuda.is_available():
data, target = data.to(device), target.to(device)
output = model(data)
test_loss += criterion(output, target).item() # mean batch loss
pred = output.data.max(1, keepdim=True)[1] # get the index of the max log-probability
correct += pred.eq(target.data.view_as(pred)).cpu().sum()
test_loss /= len(test_loader.dataset)
test_acc = round( 100. * float(correct) / len(test_loader.dataset) , 2)
if verbose :
print('Test accuracy: {}/{} ({:.2f}%)'.format(
correct, len(test_loader.dataset),
test_acc))
return test_acc, test_loss
def estimate_fisher(model, dataset, device, num = 1000, empirical = True):
# Estimate the FI-matrix for num batches of size 1
loader = torch.utils.data.DataLoader(dataset, batch_size=1, shuffle=False, num_workers=1)
est_fisher_info = {}
for n, p in model.named_parameters():
if p.requires_grad:
n = n.replace('.', '__')
est_fisher_info[n] = p.detach().clone().zero_()
model.eval()
for index,(x,y) in enumerate(loader):
if index >= num:
break
# run forward pass of model
x = x.to(device)
output = model(x)
if empirical:
# -use provided label to calculate loglikelihood --> "empirical Fisher":
label = torch.LongTensor([y]) if type(y)==int else y
label = label.to(device)
else:
label = output.max(1)[1]
negloglikelihood = torch.nn.functional.nll_loss(torch.nn.functional.log_softmax(output, dim=1), label)
model.zero_grad()
negloglikelihood.backward()
for n, p in model.named_parameters():
if p.requires_grad:
n = n.replace('.', '__')
if p.grad is not None:
est_fisher_info[n] += p.grad.detach() ** 2
est_fisher_info = {n: p/index for n, p in est_fisher_info.items()}
return est_fisher_info
def EWC_loss(model, previous_tasks_fisher, previous_tasks_parameters, current_task_index, device, ewc_lambda=5000):
if current_task_index == 0: #no task to remember -> return 0
return torch.tensor(0.).to(device)
else:
losses = []
for task_idx in range(current_task_index): # for all previous tasks and parameters
for n, p in model.named_parameters():
if ((p.requires_grad) and (n.find('bn') == -1)):
n = n.replace('.', '__')
mean = previous_tasks_parameters[n][task_idx]
fisher = previous_tasks_fisher[n][task_idx]
#print('in ewc loss, param =', p[0,0])
losses.append((fisher * (p-mean)**2).sum())
return ewc_lambda*(1./2)*sum(losses)
def update_omega(model, omega, p_prev, W, epsilon=0.1):
for n, p in model.named_parameters():
if n.find(‘bn’) == -1: # not batchnorm
if p.requires_grad:
n = n.replace(‘.’, ‘‘)
if isinstance(model, BNN):
p_current = p.org.detach().clone() # sign()
else:
p_current = p.detach().clone()
p_change = p_current - p_prev[n]
omega_add = W[n]/(p_change**2 + epsilon)
omega[n] += omega_add
print(‘parameter :\t’, n, ‘\nomega :\t’, omega[n])
W[n] = p.data.clone().zero_()
return omega
def update_W(model, W, p_old, args):
for n, p in model.named_parameters():
if p.requires_grad and (n.find(‘bn’)==-1):
n = n.replace(’.', '’)
if p.grad is not None:
if isinstance(model, BNN):
if args.bin_path:
W[n].add_(-p.grad*(p.sign().detach()-p_old[n]))
else:
W[n].add_(-p.grad*(p.org.detach()-p_old[n]))
else:
W[n].add_(-p.grad*(p.detach()-p_old[n]))
if isinstance(model, BNN):
if args.bin_path:
p_old[n] = p.sign().detach().clone()
else:
p_old[n] = p.org.detach().clone()
else:
p_old[n] = p.detach().clone()
def SI_loss(model, omega, prev_params, si_lambda):
losses =
for n, p in model.named_parameters():
if p.requires_grad and (n.find(‘bn’)==-1):
n = n.replace(‘.’, ‘__’)
if isinstance(model, BNN):
losses.append((omega[n] * (p - prev_params[n].sign())**2).sum()) #org or sign
print(‘p =\t’,p,‘\np_prev =\t’, prev_params[n])
else:
losses.append((omega[n] * (p - prev_params[n])**2).sum())
return si_lambda*sum(losses)
def switch_sign_induced_loss_increase(model, loader, bins = 10, sample = 100, layer = 2, num_run = 1, verbose = False):
model.eval() # model to evaluation mode
criterion = torch.nn.CrossEntropyLoss(reduction='none') # crossentropy loss
mbs = loader.batch_size
initial_weights = torch.empty_like(model.layers['fc'+str(layer)].weight.org).copy_(model.layers['fc'+str(layer)].weight.org)
max_magnitude = initial_weights.abs().max().item()
hidden_value_total = torch.zeros((bins, num_run,1))
total_result = torch.zeros((bins, num_run,1))
effective_bin_index = []
bins_total_candidates = []
# constructing the switch masks for every bin
for k in range(bins):
hidden_value_run = [] # will contain mean magnitude of every run
switch_list = [] # will contain a switch mask of fixed number of weight and bin for every run
for run in range(num_run):
# selecting weight candidate for switching by absolute magnitude belonging to bin
switch_indices = torch.where((initial_weights.abs() > (k/bins)*max_magnitude)*(initial_weights.abs() < ((k+1)/bins)*max_magnitude), -torch.ones_like(initial_weights), torch.ones_like(initial_weights))
bin_total = -1*switch_indices[switch_indices == -1].sum().item() # total of candidates
if run==0:
bins_total_candidates.append(bin_total)
if bin_total>=sample: # only if number of candidates greater than sample
cutoff = torch.ones_like(switch_indices[switch_indices==-1])
cutoff[sample:] *= -1 # removing candidates after accepting sample candidates
permut = torch.randperm(cutoff.nelement()) # shuffling to have different candidates every runs
switch_indices[switch_indices==-1] *= cutoff[permut] # mask with only sample candidates switch of bin k
switch_list.append(switch_indices)
effective_switch = -1*switch_indices[switch_indices==-1].sum().item()
assert(effective_switch == sample) # make sure the mask has exactly sample switches
mean_hidden_value = initial_weights[switch_indices==-1].abs().sum().item()/sample
hidden_value_run.append(mean_hidden_value/max_magnitude)
else: # rejecting bins with not enough candidates
pass
effective_run = len(hidden_value_run)
iter_per_epoch = int(len(loader.dataset)/mbs)
if effective_run>0: # in this case effective_run = num_run
effective_bin_index.append(k)
loss_total = torch.zeros((effective_run, len(loader.dataset))) # initializing result tensor
for idx, (data, target) in enumerate(loader):
if torch.cuda.is_available():
data, target = data.cuda(), target.cuda()
output_initial = model(data)
loss_initial_batch = criterion(output_initial, target) # batch loss
if (idx%(iter_per_epoch/2))==0 and verbose:
print('\nloss_initial_batch =', loss_initial_batch)
for r in range(effective_run): # loop over runs at fixed bin and batch
if idx==0 and verbose:
print('\nbin =', k, ' run =', r)
model.layers['fc'+str(layer)].weight.org.mul_(switch_list[r])
control = (model.layers['fc'+str(layer)].weight.org - initial_weights)
if idx==0 and verbose:
print('mean value of switched hidden weights (must represent the bin)=', control.abs().sum().item()/(2*sample))
output_switch = model(data)
loss_switch_batch = criterion(output_switch, target) # batch loss
control = (model.layers['fc'+str(layer)].weight - initial_weights.sign())
if idx==0 and verbose:
print('mean value of swtiched binary weight (must equal 1)=', control.abs().sum().item()/(2*sample))
print('delta_loss_batch =', (loss_switch_batch-loss_initial_batch))
model.layers['fc'+str(layer)].weight.org.mul_(switch_list[r])
control = (model.layers['fc'+str(layer)].weight.org - initial_weights)
if idx==0 and verbose:
print('delta hidden after switch back (must be zero) =', control.abs().sum().item()/(2*sample))
loss_total[r, idx*mbs:(idx+1)*mbs] = ((loss_switch_batch - loss_initial_batch)/sample).detach()
hidden_value_total[k,:] = torch.tensor(hidden_value_run).view(effective_run,1)
total_result[k,:,:] = loss_total.mean(dim=1).view(effective_run,1) # mean over training data
if verbose:
print('list of candidates per bin =', bins_total_candidates)
date = datetime.now().strftime('%Y-%m-%d')
time = datetime.now().strftime('%H-%M-%S')
path = 'results/'+date
effective_bin_index = torch.tensor(effective_bin_index)
hidden_value_cat_loss_increase = torch.cat([hidden_value_total[effective_bin_index,:,:], total_result[effective_bin_index,:,:]], dim=2)
if not(os.path.exists(path)):
os.makedirs(path)
torch.save(hidden_value_cat_loss_increase, path+'/'+time+'_switch_sign_induced_loss_increase_bins-'+str(len(effective_bin_index))+'_sample-'+str(sample)+'_layer-'+str(layer)+'_runs-'+str(num_run)+'.pt')
return hidden_value_cat_loss_increase