Hi all,
I am working on implementing a binary classification model for breast cancer histopathology data using an EfficientNet V2-S model. However, I observed that after 5-10 epochs the gradients in the SE blocks are going to 0 in the lower layers. This is observed only is SE blocks. Should this be an expected behavior of the SE layer?
The dataset used comprises of 7909 images. with 80% data used for training and 10% for testing and validation. Schedular used is reduceOnPlateau, with initial learning rate of 0.0009 and factor = 0.5. The optimzer used is and Adams optimizer with weight decay of 0.005.
I have tried decreasing and increasing the learning rate but i get the same result.
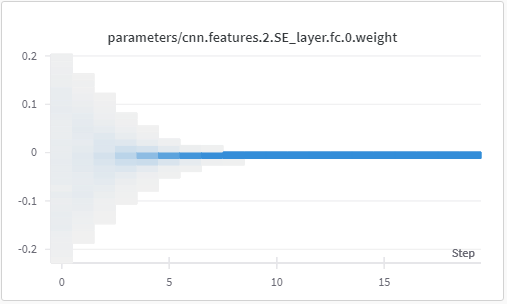
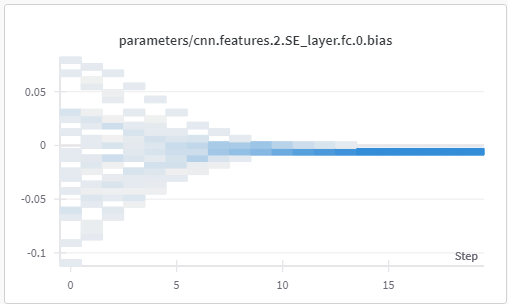
These images are of the weights and biases observed for fc layer in SE block in layer 2, for 20 epochs.
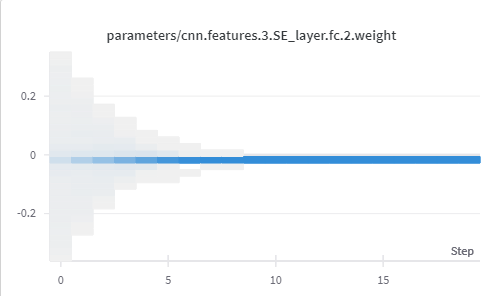

These images are of the weights and biases observed for fc layer in SE block in layer 3, for 20 epochs.
It would great if I could get some insights on why this is occurring and how can I solve this issue.
Would replacing the linear layers with 1x1 convolution make any difference?
Thanks!