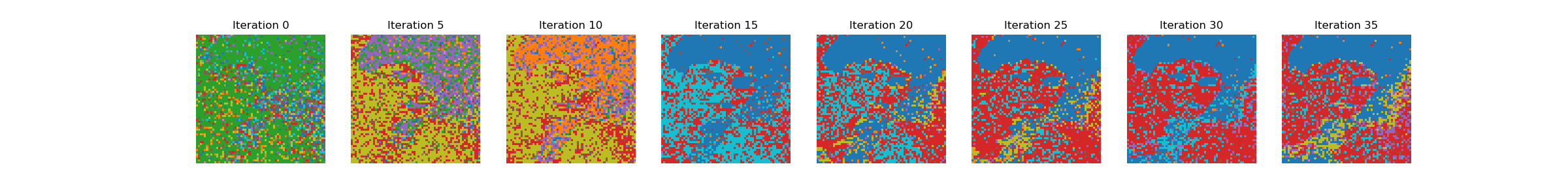
Ok, I’ve tried to boil the code down to a minimal example. Let me know if anything is unclear. Thanks again.
The code is below, but can also be found in this Gist: https://gist.github.com/isaacwasserman/ae69cfd82fc89b9083720605bcb70e93
import torch
import numpy as np
import time
from PIL import Image
import skimage.segmentation
import skimage.color
import matplotlib.pyplot as plt
# Find fastest device available
device = "cpu"
if torch.cuda.is_available():
device = "cuda"
elif torch.backends.mps.is_available():
device = "mps"
def initial_labels(image, d, n_segments, compactness=10, sigma=1):
"""Generates patch-level labels for an image using SLIC
Args:
image (ndarray): image represented by (H,W,C) array
d (int): number of patches per dimension to assign labels to; function will return d^2 labels
n_segments (int): number of segments to generate
compactness (int, optional): compactness parameter passed to SLIC. Defaults to 10.
sigma (int, optional): sigma parameter passed to SLIC. Defaults to 1.
Returns:
tensor: softmaxed labels for each patch in the image (d^2, n_segments)
ndarray: the full resolution segmentation
"""
seg = skimage.segmentation.slic(image,
n_segments=n_segments, compactness=compactness, sigma=sigma,
enforce_connectivity=False, convert2lab=True)
while len(np.unique(seg)) > n_segments:
# count number of pixels in each segment
segments, counts = np.unique(seg, return_counts=True)
# find smallest segment
smallest1 = segments[np.argmin(counts)]
# find second smallest segment
counts[smallest1] = np.max(counts)
smallest2 = segments[np.argmin(counts)]
# merge smallest segments
seg[seg == smallest1] = smallest2
t = torch.tensor(seg).unsqueeze(0).unsqueeze(0).float()
# bin the image
kernel_width = image.shape[0] // d
kernel_height = image.shape[1] // d
regions = torch.nn.functional.unfold(t, (kernel_width, kernel_height), stride=(kernel_width, kernel_height), padding=0)
regions = regions.permute(0,2,1).squeeze().to(torch.int64).squeeze(0)
# count occurences of each segment in each bin
labels = torch.nn.functional.one_hot(regions, n_segments).float()
labels = torch.sum(labels, dim=1)
labels = torch.nn.functional.softmax(labels, dim=1)
return labels, seg
class GNEMNet(torch.nn.Module):
def __init__(self, use_padding=True, patch_size=(32,32), k=10, n_filters=16, dropout=0.2):
"""FCN architecture which operates on patches rather than the entire image
Args:
patch_size (tuple, optional): shape of input patches. Defaults to (32,32).
k (int, optional): number of output channels (segments). Defaults to 10.
n_filters (int, optional): number of filters/channels in the middle of the network. Defaults to 16.
dropout (float, optional): amount of dropout during training. Defaults to 0.2.
"""
super(GNEMNet, self).__init__()
self.k = k
self.n_input_channels = 3
self.padding = 1 if use_padding else 0
padding_compensation = -4 + 4 * self.padding
self.conv1 = torch.nn.Conv2d(self.n_input_channels, n_filters, 3, padding=self.padding)
self.BN1 = torch.nn.BatchNorm2d(n_filters)
self.dropout1 = torch.nn.Dropout(dropout)
self.conv2 = torch.nn.Conv2d(n_filters, 1, 3, padding=self.padding)
self.BN2 = torch.nn.BatchNorm2d(1)
self.dropout2 = torch.nn.Dropout(dropout)
self.output = torch.nn.Conv2d(1, k, (patch_size[0] + padding_compensation, patch_size[1] + padding_compensation), k)
self.tile_size = patch_size
self.train_indices = None
self.use_subset = True
self.unfold_stride = 1
self.make_patches = True
def forward(self, x):
x = self.conv1(x)
x = self.BN1(x)
x = torch.nn.functional.tanh(x)
x = self.dropout1(x)
x = self.conv2(x)
x = self.BN2(x)
x = torch.nn.functional.tanh(x)
x = self.dropout2(x)
x = self.output(x)
return x
class PatchDL():
def __init__(self, image_tensor, initial_labels, d, batch_size):
"""Dataloader for image patches
Args:
image_tensor (tensor): input image represented by a (1, C, H, W) tensor
initial_labels (tensor): initial labels for each trainining patch represented by a (d^2, n_segments) tensor
d (_type_): _description_
batch_size (_type_): _description_
"""
self.image_tensor = image_tensor
self.labels = initial_labels
self.d = d
self.patch_size = (image_tensor.shape[2] // d, image_tensor.shape[3] // d)
self.patches = torch.nn.functional.unfold(self.image_tensor, kernel_size=self.patch_size, stride=1, dilation=1, padding=0)
self.patches = self.patches.view(self.image_tensor.shape[1], self.patch_size[0], self.patch_size[1], -1).permute(3, 0, 1, 2).to(device)
self.train_indices = torch.cat([torch.arange(self.d) * self.patch_size[0] + ((self.image_tensor.shape[2] - (self.patch_size[0] - 1)) * self.patch_size[1] * row) for row in range(d)])
self.train_indices = torch.stack([self.train_indices, torch.arange(self.d * self.d)], dim=1)
self.batch_size = int(batch_size * len(self.train_indices))
def get_train_batch(self):
"""Returns a random batch of patches with their corresponding labels from the "training set". The training set is a subset of the d^2 patches in the image that don't overlap.
Returns:
tensor: batch of patches represented by a (batch_size, C, H, W) tensor
tensor: batch of labels represented by a (batch_size, n_segments) tensor
"""
batch_indices = self.train_indices[torch.randperm(self.train_indices.shape[0])[:self.batch_size]]
patches = self.patches[batch_indices[:, 0]]
labels = self.labels[batch_indices[:, 1]]
return patches, labels
def get_inference_set(self):
"""Returns all patches in the image
Returns:
tensor: all patches represented by a (d^2, C, H, W) tensor
"""
patches = self.patches
return patches
class GNEMS_Segmentor:
def __init__(self, use_padding=True, d=16, n_filters=16, dropout=0.2, lr=0.001, subset_size=0.5, sigma=1, compactness=0.1, k=18, epochs=40):
"""Segmentor class
Args:
use_padding (bool, optional): whether or not to use padding in the CNN. Defaults to True.
d (int, optional): square root of the number of patches to divide the image into. Defaults to 16.
n_filters (int, optional): number of filters/channels in the middle of the CNN. Defaults to 16.
dropout (float, optional): amount of dropout used during training. Defaults to 0.2.
lr (float, optional): learning rate for network. Defaults to 0.001.
subset_size (float, optional): percentage of training set to use in each batch. Defaults to 0.5.
sigma (int, optional): sigma parameter passed to SLIC. Defaults to 1.
compactness (float, optional): compactness parameter passed to SLIC. Defaults to 0.1.
k (int, optional): number of segments to divide the image into. Defaults to 18.
epochs (int, optional): number of times the training loop should run. Defaults to 40.
"""
self.d = d
self.n_filters = n_filters
self.dropout = dropout
self.lr = lr
self.subset_size = subset_size
self.net = None
self.slic_segments = k
self.sigma = sigma
self.compactness = compactness
self.k = k
self.initial_labels = None
self.epochs = epochs
self.initial_segmentation = None
self.intermediate_cross_entropies = []
self.image_size = None
self.patch_size = None
self.use_padding = use_padding
def fit(self, image):
"""Fits the network to the image
In this minimal example, the fit method standardizes the image, generates labels using `inital_labels()` (SLIC), and trains the network in a supervised manner, using cross entropy loss.
Args:
image (ndarray): input image represented by a (H,W,C) array
"""
self.image_tensor = torch.tensor(skimage.color.rgb2lab(image), dtype=torch.float32).to(device).permute(2, 0, 1).unsqueeze(0)
self.image_size = self.image_tensor.shape[-2:]
self.patch_size = (self.image_size[0] // self.d, self.image_size[1] // self.d)
# standardize image to [-1,1]
cur_min = self.image_tensor.min()
cur_max = self.image_tensor.max()
self.image_tensor = (2 * (self.image_tensor - cur_min)/(cur_max - cur_min)) - 1
# create labels using SLIC
self.initial_labels, self.initial_segmentation = initial_labels(image, self.d, self.k, sigma=self.sigma, compactness=self.compactness)
self.initial_labels = self.initial_labels.argmax(dim=1).to(device)
# create dataloader
self.dataloader = PatchDL(self.image_tensor, self.initial_labels, self.d, self.subset_size)
# Initialize CNN
self.net = GNEMNet(use_padding=self.use_padding, patch_size=self.patch_size, n_filters=self.n_filters, dropout=self.dropout, k=self.k).to(device)
self.net.train()
# Initialize optimizer and loss function
cross_entropy = torch.nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(self.net.parameters(), lr=self.lr)
# Train CNN
for epoch in range(self.epochs):
patches, labels = self.dataloader.get_train_batch()
optimizer.zero_grad()
outputs = self.net(patches).squeeze(-1).squeeze(-1)
loss = cross_entropy(outputs, labels)
self.intermediate_cross_entropies.append(loss.item())
loss.backward()
optimizer.step()
def predict(self):
"""Predicts the segmentation of the image
Runs all possible patches in the image through the network, forms the output into a single image, uses bilinear interpolation to upscale the output to the original image size, and returns the greatest channel index for each pixel.
Returns:
ndarray: segmentation represented by a (H,W) array
"""
self.net.eval()
patches = self.dataloader.get_inference_set()
collage_width = np.sqrt(patches.shape[0]).astype(int)
# Get predictions for each patch
outputs = self.net(patches).detach().squeeze(-1).squeeze(-1).unsqueeze(0)
# Reshape predictions into an image
outputs = outputs.permute(0, 2, 1)
outputs = outputs.reshape(1, self.k, collage_width, collage_width)
# Upscale the image slightly to the original image size
outputs = torch.nn.functional.interpolate(outputs, self.image_tensor.shape[2:])
# Return greatest channel index for each pixel
outputs = outputs.argmax(1).squeeze(0).cpu().numpy()
return outputs
# Get an image
image_url = "https://hips.hearstapps.com/hmg-prod/images/dog-puppy-on-garden-royalty-free-image-1586966191.jpg"
!wget -nc {image_url}
image_path = image_url.split("/")[-1]
image = np.array(Image.open(image_path).resize((512, 512)))[:,:,:3]
# Set hyperparameters
k = 18 # number of segments
d = 64 # number of patches across image (number of patches = d^2)
lr = 0.01 # learning rate
subset_size = 0.5 # batch size as a fraction of total number of patches
epochs = 40 # number of epochs per iteration
USE_PADDING = True # whether to use padding in the CNN
# Initialize and fit segmentor
segmentor = GNEMS_Segmentor(use_padding=USE_PADDING, k=k, d=d, subset_size=subset_size, lr=lr, epochs=epochs, n_filters=16, compactness=0.01, sigma=1)
segmentor.fit(image)
# Predict segmentation
seg = segmentor.predict()
# Plot loss curve
plt.plot(segmentor.intermediate_cross_entropies)
plt.legend(["Cross-entropy"])
plt.show()
# Plot input image
plt.imshow(image)
plt.title("Input Image")
plt.show()
# Plot initial segmentation by SLIC
plt.imshow(segmentor.initial_segmentation, cmap="tab10")
plt.title("Initial Segmentation")
plt.show()
# Plot initial labels (downsampled SLIC segmentation)
plt.imshow(segmentor.initial_labels.to("cpu").numpy().reshape(d,d), cmap="tab10")
plt.title("Initial Patch Labels")
plt.show()
# Plot predicted segmentation
plt.imshow(seg, cmap="tab10")
plt.title("Prediction")
plt.show()