Dear Altruists,
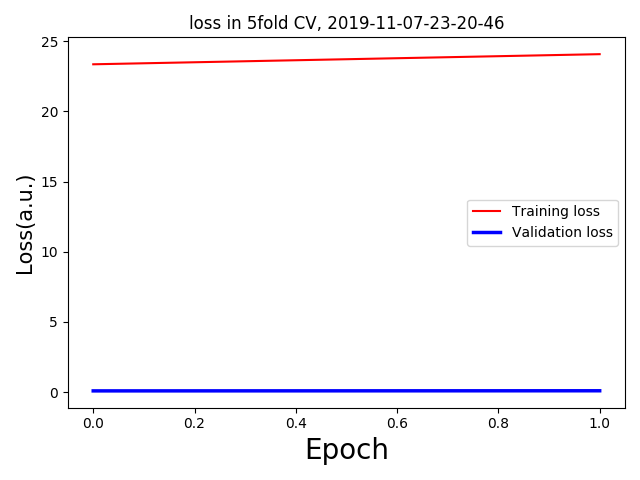
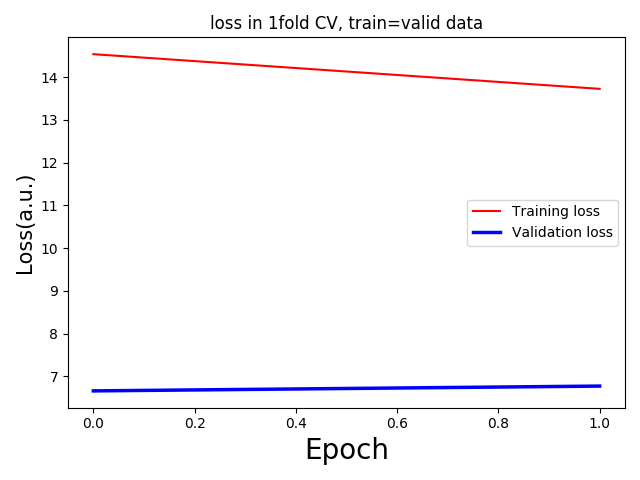
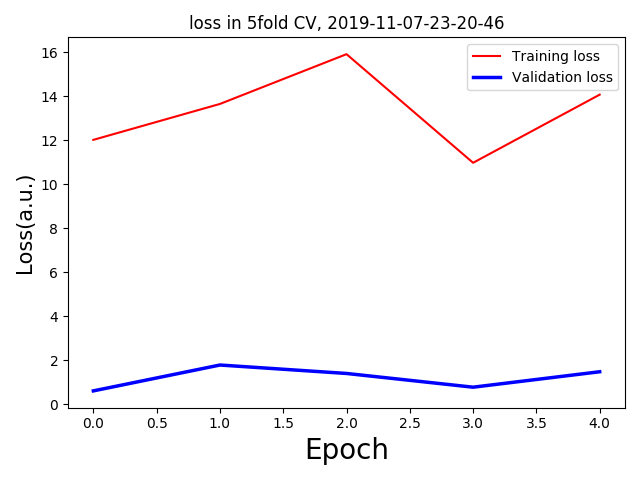
I am running some regression analysis with 3D MRI data. But I am getting too low validation loss with respect to the training loss. For 5 fold validation, each having only one epoch(as a trial) I am getting the following loss curves:
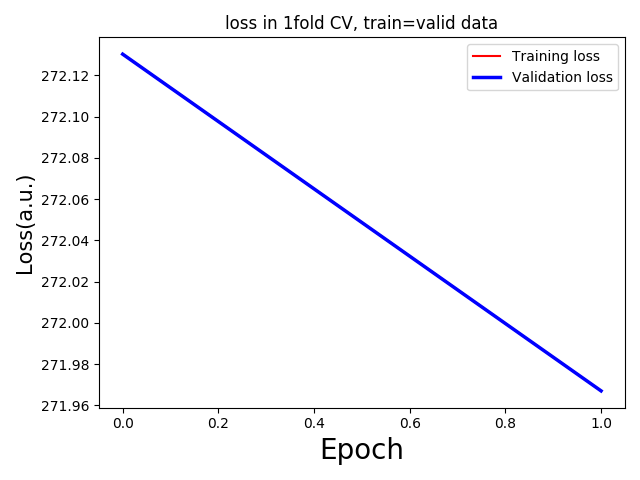
To debug the issue, I used the same input and target for training and validation setups in my codes.
For building model:
def build_model():
torch.cuda.manual_seed(1)
model = ResNet3D().to(device)
model.apply(weights_init)
Optimizer = optim.Adam(model.parameters(), lr=lr)
Criterion = nn.MSELoss().cuda()
return model, Optimizer, Criterion
I will run the build_model function for every fold that is my plan.
Net, Optimizer, Criterion = build_model()
train_loader = data.DataLoader(train_dataset, batch_size=batchsize, shuffle=True) #for every cross-validation fold train_dataset changes.
Training section:
Net.train()
sample = train_loader.dataset[0]
input = sample[0] #just taking one sample for loss calculation
input = input.unsqueeze(0).float().to(device) #torch.Size([1, 86, 110, 78])
target = sample[1]
target = target.unsqueeze(0).float().to(device) #torch.Size([1, 256])
output = Net(input) #torch.Size([1, 256])
Optimizer.zero_grad()
loss = Criterion(output,target)
print("Loss in training:",loss.item())
Loss in training: 14.414422035217285
** I did not update Net weights Optimizer.step() or neither did back propagation loss.backward()
Validation section:
Net.eval()
with torch.no_grad():
outputvalid = Net(input) #same input in same Net
lossvalid = Criterion(outputvalid,target)
print("Loss in validation: ",lossvalid.item())
Loss in validation: 2.8760955333709717
What could be the reason??