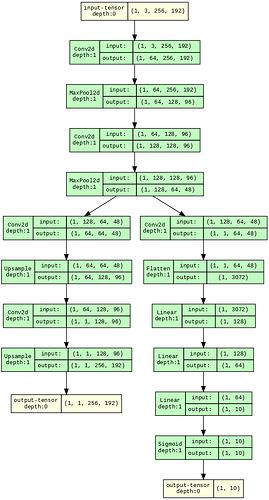
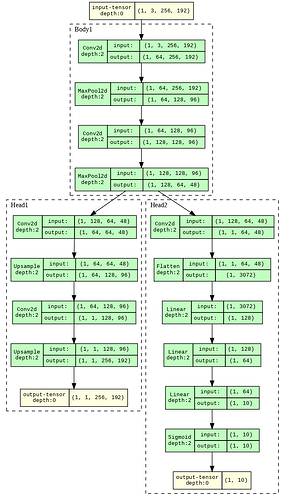
I am creating two models using the same stack of layers using a single functional class and another with discrete classes, later accumulating into one class. Even model.summary() gives the same number of parameters for both models; why does the size of Forward/backward pass (618.00 MB vs. 729.38 MB) differ by some amount using torch summary? Even the training results are inconsistent, using the same seed for both the models and every hyper-parameter the same.
Could you post both model architectures and did you measure the actual memory usage yourself or only via torchsummary?
I measured the memory using torchsummary. I am showing an example (since the actual model was too big) here (Note the difference is small here); I want to know what exactly is creating the difference (56.39 vs 53.02); also, the results were inconsistent in the model that I was using during training by just using the class-based method even when using proper seed in everything…
! pip install -q torchview
! pip install -q -U graphviz
import torch
import torch.nn as nn
from torch import Tensor
import torch.nn.functional as F
from torchsummary import summary
from torchsummary import summary
from torchview import draw_graph
from torchvision.models import resnet18, GoogLeNet, densenet, vit_b_16
# when running on VSCode run the below command
# svg format on vscode does not give desired result
import graphviz
graphviz.set_jupyter_format('png')
import matplotlib.pyplot as plt
use_cuda = torch.cuda.is_available()
print('use_cuda: {}'.format(use_cuda))
device = torch.device("cuda" if use_cuda else "cpu")
print("Device to be used : ",device)
import torch
import torch.nn as nn
import torch.nn.functional as F
from torchsummary import summary
class Test_Model_1(nn.Module):
def __init__(self, inp_ch):
super().__init__()
self.conv1 = nn.Conv2d(inp_ch, 64, 3, padding=1)
self.conv2 = nn.Conv2d(64, 128, 3, padding=1)
self.conv3 = nn.Conv2d(128, 64, 3, padding=1)
self.conv4 = nn.Conv2d(64, 1, 3, padding=1)
self.upsample = nn.Upsample(scale_factor=2, mode='bilinear', align_corners=True)
self.conv_before_flatten = nn.Conv2d(128, 1, 1)
self.dropout = nn.Dropout(p=0.2)
self.maxpool = nn.MaxPool2d(2)
self.flatten = nn.Flatten()
self.fc1 = nn.Linear(3072, 128)
self.fc2 = nn.Linear(128, 64)
self.fc3 = nn.Linear(64, 10)
self.sigmoid = nn.Sigmoid()
def forward(self, x):
conv1 = self.conv1(x)
x = self.maxpool(conv1)
conv2 = self.conv2(x)
x = self.maxpool(conv2)
conv3 = self.upsample(self.conv3(x))
conv4 = self.upsample(self.conv4(conv3))
conv_flatten = self.conv_before_flatten(x)
flatten = self.flatten(conv_flatten)
fcns = self.fc3(self.fc2(self.fc1(flatten)))
out = self.sigmoid(fcns)
return out, conv4
model1 = Test_Model_1(inp_ch=3)
model1 = model.to(device)
summary(model1, input_size=(3, 256, 192))
##### Here is the assembled class based implementation
import torch
import torch.nn as nn
import torch.nn.functional as F
from torchsummary import summary
class Body1(nn.Module):
def __init__(self, inp_ch):
super().__init__()
self.conv1 = nn.Conv2d(inp_ch, 64, 3, padding=1)
self.conv2 = nn.Conv2d(64, 128, 3, padding=1)
self.maxpool = nn.MaxPool2d(2)
def forward(self, x):
conv1 = self.conv1(x)
x = self.maxpool(conv1)
conv2 = self.conv2(x)
x = self.maxpool(conv2)
return x
class Head1(nn.Module):
def __init__(self):
super().__init__()
self.conv3 = nn.Conv2d(128, 64, 3, padding=1)
self.conv4 = nn.Conv2d(64, 1, 3, padding=1)
self.upsample = nn.Upsample(scale_factor=2, mode='bilinear', align_corners=True)
def forward(self, x):
conv3 = self.upsample(self.conv3(x))
conv4 = self.upsample(self.conv4(conv3))
return conv4
class Head2(nn.Module):
def __init__(self):
super().__init__()
self.conv_before_flatten = nn.Conv2d(128, 1, 1)
self.dropout = nn.Dropout(p=0.2)
self.flatten = nn.Flatten()
self.fc1 = nn.Linear(3072, 128)
self.fc2 = nn.Linear(128, 64)
self.fc3 = nn.Linear(64, 10)
self.sigmoid = nn.Sigmoid()
def forward(self, x):
conv_flatten = self.conv_before_flatten(x)
flatten = self.flatten(conv_flatten)
fcns = self.fc3(self.fc2(self.fc1(flatten)))
out = self.sigmoid(fcns)
return out
class Test_Model_2(nn.Module):
def __init__(self, inp_ch):
super().__init__()
self.body1 = Body1(inp_ch)
self.head1 = Head1()
self.head2 = Head2()
def forward(self, x):
body_1 = self.body1(x)
head_1 = self.head1(body_1)
head_2 = self.head2(body_1)
return head_1, head_2
model2 = Test_Model_2(inp_ch=3)
model2 = model.to(device)
summary(model2, input_size=(3, 256, 192))
model1.summary() gives the following:
----------------------------------------------------------------
Layer (type) Output Shape Param #
================================================================
Conv2d-1 [-1, 64, 256, 192] 1,792
MaxPool2d-2 [-1, 64, 128, 96] 0
Conv2d-3 [-1, 128, 128, 96] 73,856
MaxPool2d-4 [-1, 128, 64, 48] 0
Conv2d-5 [-1, 64, 64, 48] 73,792
Upsample-6 [-1, 64, 128, 96] 0
Conv2d-7 [-1, 1, 128, 96] 577
Upsample-8 [-1, 1, 256, 192] 0
Conv2d-9 [-1, 1, 64, 48] 129
Flatten-10 [-1, 3072] 0
Linear-11 [-1, 128] 393,344
Linear-12 [-1, 64] 8,256
Linear-13 [-1, 10] 650
Sigmoid-14 [-1, 10] 0
================================================================
Total params: 552,396
Trainable params: 552,396
Non-trainable params: 0
----------------------------------------------------------------
Input size (MB): 0.56
Forward/backward pass size (MB): 53.02
Params size (MB): 2.11
Estimated Total Size (MB): 55.69
----------------------------------------------------------------
While, model2.summary() gives:
----------------------------------------------------------------
Layer (type) Output Shape Param #
================================================================
Conv2d-1 [-1, 64, 256, 192] 1,792
MaxPool2d-2 [-1, 64, 128, 96] 0
Conv2d-3 [-1, 128, 128, 96] 73,856
MaxPool2d-4 [-1, 128, 64, 48] 0
Body1-5 [-1, 128, 64, 48] 0
Conv2d-6 [-1, 64, 64, 48] 73,792
Upsample-7 [-1, 64, 128, 96] 0
Conv2d-8 [-1, 1, 128, 96] 577
Upsample-9 [-1, 1, 256, 192] 0
Head1-10 [-1, 1, 256, 192] 0
Conv2d-11 [-1, 1, 64, 48] 129
Flatten-12 [-1, 3072] 0
Linear-13 [-1, 128] 393,344
Linear-14 [-1, 64] 8,256
Linear-15 [-1, 10] 650
Sigmoid-16 [-1, 10] 0
Head2-17 [-1, 10] 0
================================================================
Total params: 552,396
Trainable params: 552,396
Non-trainable params: 0
----------------------------------------------------------------
Input size (MB): 0.56
Forward/backward pass size (MB): 56.39
Params size (MB): 2.11
Estimated Total Size (MB): 59.06
----------------------------------------------------------------
Even the layers are the same when seeing it visually…
So, why does class-based implementation increase the Forward/backward pass size?
The primary motivation for creating the class-based discrete blocks is I want to set require_grad=False/True in the entire class (or block, like head1 or head2 like a switch, and set require_grad=True, in different stages of training), and I don’t have to do that iteratively in the first implementation. @ptrblck Any suggestions on how to set the block’s required grad=True and False, in the first model implementation, without going through each layer’s bias and weights individually in the first (non-discrete class-based) implementation?
The difference in memory is most likely coming from the different allocations of intermediate activations in the forward method. You would have to make sure to assign the same number of outputs to variables or reassign them.
Seeding can be tricky as you would need to guarantee the exact same order or calls is made to the pseudorandom number generator. I’m not on my workstation now but you could debug it already by checking the parameters layer by layer.
Having the same issue, did you figure out why it is larger when using more nn.Modules? In my case, I created the following network:
def __init__():
layer0 = SomeCustomizedModule1
layer1 = SomeCustimizedModule2
def foward(self, x):
x = layer0(x)
x = layer1(x)
return x
and if I move layer1 into SomeCustomizedModule1, the forward/backward passing size dramatically drops. Really weird : (
Would you post the definitions of all used classes?
Hi, I figured out why the sizes differ. The forward/backward passing size is calculated by adding all the modules’ output sizes together.
Say we have 2 convs in our network:
def __init__(self):
self.conv1 = nn.Conv2d(3, 64, 3, padding=1)
self.conv2 = nn.Conv2d(64, 128, 3, padding=1)
def forward(self, x):
x = self.conv1(x) --> output1
x = self.conv2(x) --> output2
return x
The f/bp size would be output1 size + output2 size.
If we wrap them into one nn.Module, for example:
class XXX(nn.Module):
def __init__(self):
self.conv1 = ...
self.conv2 = ...
def forward(self, x):
x = self.conv1(x) --> output1
x = self.conv2(x) --> output2
return x
-------------------
def __init__(self):
self.module = XXX()
def forward(self, x):
x = self.module(x) --> output3
return x
The final f/bp size would be output1 + output2 + output3!
There would be a redundant output for each wrapping Module!
Would this be a torchsummary bug?
No idea; I couldn’t figure this thing out. Even the training had a minor difference in optimization when using proper seeding.
I don’t fully understand the explanation as calling self.module(x) will just return the same output from self.conv2, which you previously tagged as output2.
Also, what does final f/bp size would be output1 + output2 + output3 mean in this context? Shapes are not added together so I guess you are using a different syntax to describe intermediate shapes?