Hello,
Sorry if I dont give all the info from the start, this is my first post on forums.
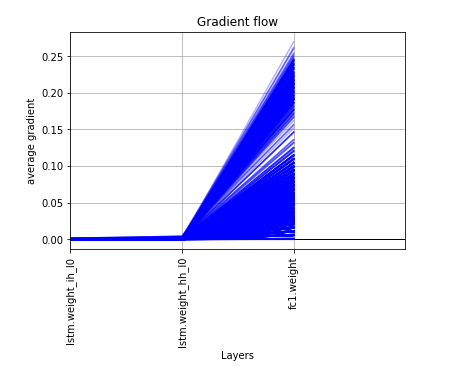
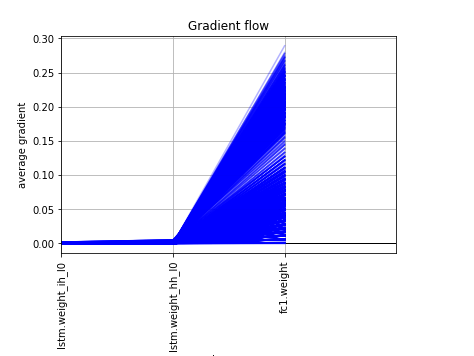
I’ve been using as a feature extractor from text Word2Vec from gensim to have word embedding and at training with a pytorch model the validation result are very alterning from epoch to epoch, I think due to the gradients being very concentrated in the last layer and close to 0 to the other layers. I attached also a plot from the layers gradients. The thing that bothered me is that scikit-learn MLPRegressor gives more stable results out of the same features and replicated in pytorch, giving different results.
The model looks like this, using L1Loss and Adam optimizer
class Net(nn.Module):
Preformatted textdef __init__(self):
super(Net, self).__init__()
self.h0 = torch.randn(1, 1, 150).cuda()
self.c0 = torch.randn(1, 1, 150).cuda()
self.lstm = nn.LSTM(200,150,1)
for name, param in self.lstm.named_parameters():
if 'bias' in name:
pass
elif 'weight' in name:
nn.init.xavier_normal_(param,gain=nn.init.calculate_gain('tanh'))
self.fc1 = nn.Linear(150,500)
nn.init.xavier_normal_(self.fc1.weight,gain=nn.init.calculate_gain('tanh'))
self.fc2 = nn.Linear(500,300)
nn.init.xavier_normal_(self.fc2.weight,gain=nn.init.calculate_gain('tanh'))
self.fc3 = nn.Linear(300, 2)
nn.init.xavier_normal_(self.fc3.weight,gain=nn.init.calculate_gain('tanh'))
#self.bc1 = nn.BatchNorm1d(150)
#self.bc3 = nn.LayerNorm(300)
self.f = nn.Tanh()
def forward(self, x):
x = x.unsqueeze(1)
x,(self.h0,self.c0) = (self.lstm(x,(self.h0,self.c0)))
x = self.f(x)
#print(x.shape)
x = x.flatten(1)
x = self.f(self.fc1(x))
x = self.f(self.fc2(x))
x = self.fc3(x)
return x
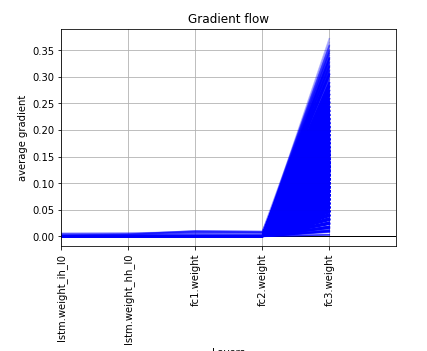
I want to know why this happens and why I can’t achieve better results then scikit-learn MLPRegressor