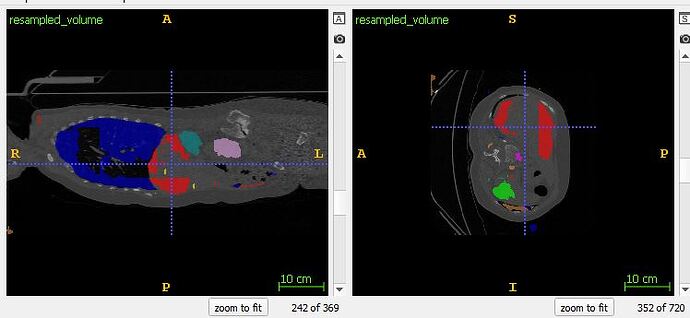
Hi All,
I’m having some issues using a 3D UNet (base 32, depth 4) for multi-organ segmentation. Due to memory constraints, I use 128x128,128 patches with a sliding window, with overlap of 32 in each axis. I use a combined loss of weighted DICE and weighted CE, an adam optimizer with lr=0.00001. The network has learned something, and results (as you’ll see) look…interesting.
The following inference images come from a similar patch-based approach. To avoid discontinuities at the edges, I sample the volume with a sliding window larger than the area of interested, and crop the 128x128x128 output that I’m interested in from the output of this larger sliding window. In this way, each sample contains more context from the volume to smooth the output between subsequent samples.
Despite this approach, I’m still finding discontinuities at the edges of each sample. Note: I grab the 128x128x128 center of each larger sliding window. The following images come from the output of coronal slice 128, and coronal slice 129: which is the border between subsequent samples.
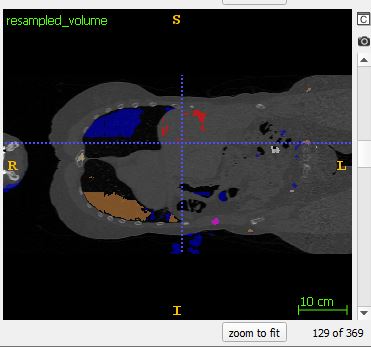
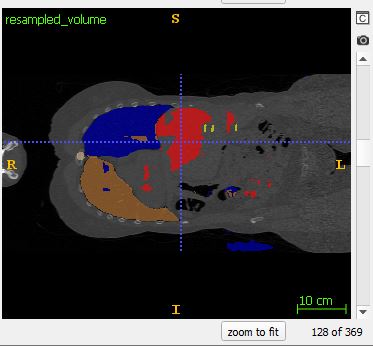
Moreover, blocky behavior (Also noted here, but unfortunately never answered: How to avoid 'blocky' output when stitching 3D patches back together?)
can also be witnessed in other slices. Here particularly in the lung and liver segmentations :
Anyone have experience with this problem that can give some advice would be greatly appreciated!