Hi,
I have volumes of size [120*768*768] which I have broken down into patches of size [32*256*256] to fit on GPU.
for prediction of a full volume the patches are stitched back together to get the full output. I’m using the code shared here (https://github.com/wolny/pytorch-3dunet/blob/master/predict.py#L97) for stitching the patches back into full size.
Basically what I’m doing is initializing a prediction_map of size [120*768*768] and a normalization_mask of the same size.
# initialize the output prediction arrays
prediction_maps = [np.zeros(prediction_maps_shape, dtype='float32') ]
# initialize normalization mask in order to average out probabilities of overlapping patches
normalization_masks = [np.zeros(prediction_maps_shape, dtype='float32')]
Then probabilities of each patch by network model is aggregated into this at the positions given by u_index.
# accumulate probabilities into the output prediction array
prediction_map[u_index] += u_prediction
# count voxel visits for normalization
normalization_mask[u_index] += 1
index is returned by data loader which keeps track of the patch slices.
The author of the code uses an unpad script to prevent the very thing that is happening with my output here:
# unpad in order to avoid block artifacts in the output probability maps
u_prediction, u_index = utils.unpad(prediction, index, volume_shape)
https://github.com/wolny/pytorch-3dunet/blob/master/unet3d/utils.py#L121
In the end the aggregated into prediction_map is divided by the normalization_ask to average the output.
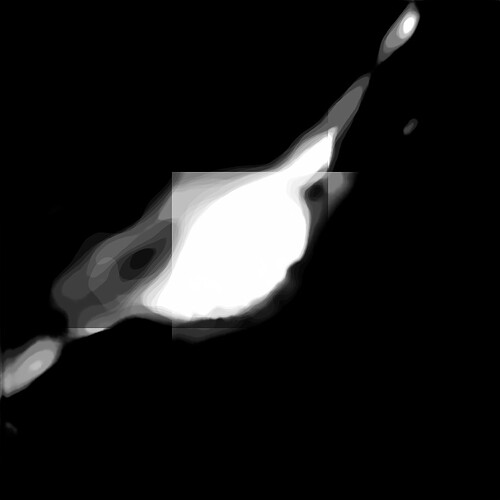
This whole process should not show a blocky output but still I get a result like this for patch size [32, 512, 512] and stride [2, 256, 256]. There is considerable overlap and as per author’s guidelines this should not cause block artefacts.
The network is working perfectly for individual patches (Ive checked from TensorBoardX) but its failing at this stitching stage.
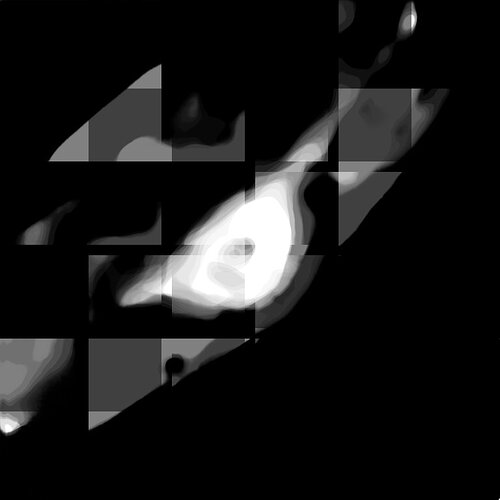
And when I use a patch size of [32, 256, 256] and stride [2, 128, 128], I get the following strange output:
What am I doing wrong? Is there any better way to stitch these patches back together? Would torch.unfold and torch.fold help me here?
Some thoughts/ help would be appreciated
Thank you