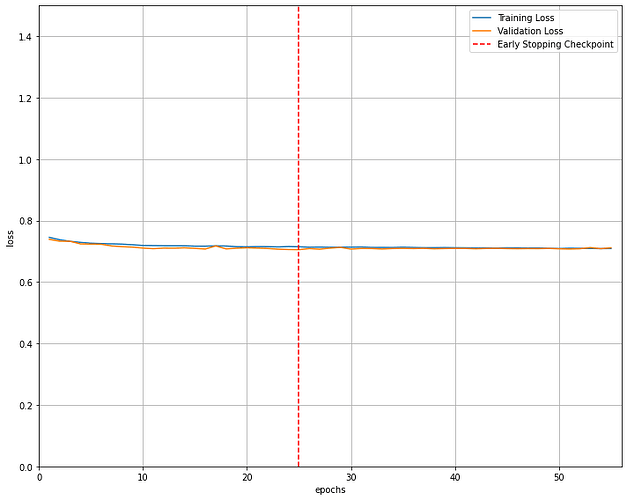
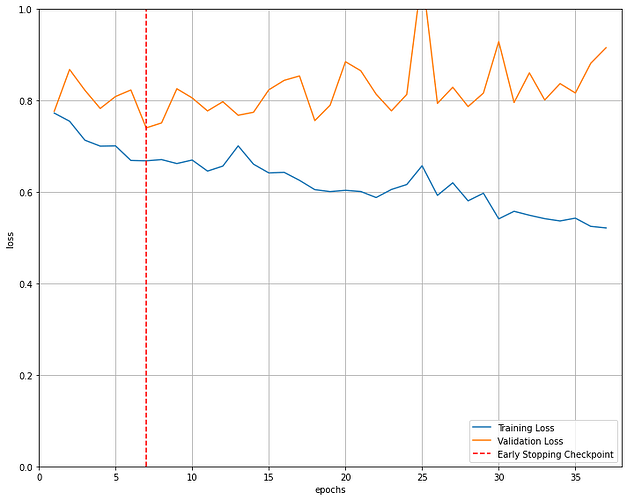
Thank you very much. I get the following error:
RuntimeError: Given groups=1, weight of size [32, 1, 1, 10], expected input[1, 6000, 1, 301] to have 1 channels, but got 6000 channels instead
My model is:
class ConvNet1D(nn.Module):
def init(self):
super(ConvNet1D,self).init()
self.features = nn.Sequential(
nn.Conv2d(1, 32, kernel_size=(1,10), stride=1, padding=0), # change this
nn.ReLU(inplace=True),
)
self.flat = nn.Flatten()
#ialize our softmax classifier
self.fc2 = nn.Linear(in_features=3213504, out_features=2) #classes =2
self.logSoftmax = nn.LogSoftmax(dim=1)
def forward(self, input1, input2,input3,input4,input5,input6,input7,input8,input9,input10,input11,input12):
xe1 = self.features(input1)
xe2= self.features(input2)
xe3 = self.features(input3)
xe4 = self.features(input4)
xe5 = self.features(input5)
xe6 = self.features(input6)
xe7 = self.features(input7)
xe8 = self.features(input8)
xe9 = self.features(input9)
xe10 = self.features(input10)
xe11 = self.features(input11)
xe12 = self.features(input12)
f1=self.flat(xe1)
f2=self.flat(xe2)
f3=self.flat(xe3)
f4=self.flat(xe4)
f5=self.flat(xe5)
f6=self.flat(xe6)
f7=self.flat(xe7)
f8=self.flat(xe8)
f9=self.flat(xe9)
f10=self.flat(xe10)
f11=self.flat(xe11)
f12=self.flat(xe12)
#con = torch.cat((f1), 1) #f2,f3,f4,f5,f6,f7,f8,f9,f10,f11,f12
combined = torch.cat((f1.view(f1.size(0), -1),
f2.view(f2.size(0), -1),f3.view(f1.size(0), -1),
f4.view(f1.size(0), -1),f5.view(f1.size(0), -1),
f6.view(f1.size(0), -1),f7.view(f1.size(0), -1),
f8.view(f1.size(0), -1),f9.view(f1.size(0), -1),
f10.view(f1.size(0), -1),f11.view(f1.size(0), -1),f12.view(f1.size(0), -1),), dim=1)
f3=self.flat(combined)
fc=self.fc2(f3)
x=self.logSoftmax(fc)
return x
and this is how I train
model.train()
trainECG1=trainECG1.to(device)
trainECG2=trainECG2.to(device)
trainECG3=trainECG3.to(device)
trainECG4=trainECG4.to(device)
trainECG5=trainECG5.to(device)
trainECG6=trainECG6.to(device)
trainECG7=trainECG7.to(device)
trainECG8=trainECG8.to(device)
trainECG9=trainECG9.to(device)
trainECG10=trainECG10.to(device)
trainECG11=trainECG11.to(device)
trainECG12=trainECG12.to(device)
trainY=trainY.to(device)
valY=valY.to(device)
valECG1 = valECG1.to(device)
valECG2 = valECG2.to(device)
valECG3 = valECG3.to(device)
valECG4 = valECG4.to(device)
valECG5 = valECG5.to(device)
valECG6 = valECG6.to(device)
valECG7 = valECG7.to(device)
valECG8 = valECG8.to(device)
valECG9 = valECG9.to(device)
valECG10 = valECG10.to(device)
valECG11 = valECG11.to(device)
valECG12 = valECG12.to(device)
optimizer.zero_grad()
y_train_pred = model(trainECG1,trainECG2,trainECG3,trainECG4,trainECG5,trainECG6,trainECG7,trainECG8,
trainECG9,trainECG10,trainECG11,trainECG12)
train_loss = criterion(y_train_pred, trainY.unsqueeze(1))
My inputs size are 60001301 each. I mean, trainECG1= 60001301, trainECG2=60001301,etc.
No. of samples = 6000, I have 1* 301 time series ECG signal. My output is binary classification.
Can you please help me to resolve the error?
Thank you very much.