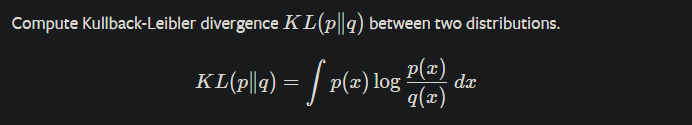I have two multivariate Gaussian distributions that I would like to calculate the kl divergence between them. each is defined with a vector of mu and a vector of variance (similar to VAE mu and sigma layer). What is the best way to calculate the KL between the two? Is this even doable? because I do not have the covariance matrix. If that is not doable, what if I take samples from both distributions and calculate the KL between the samplings? is that the right way of doing this? is there any build-in function for any of these calculations?
Hi,
Yes, this is the correct approach.
Just be aware that the input a must should contain log-probabilities and the target b should contain probability.
https://pytorch.org/docs/stable/nn.functional.html?highlight=kl_div#kl-div
By the way, PyTorch use this approach:
https://pytorch.org/docs/stable/distributions.html?highlight=kl_div#torch.distributions.kl.kl_divergence
Good luck
Nik
Thanks Nick for your input. I should restate my question. I have two multi-variate distributions each defined with “n” mu and sigma. Now I would like to do a KL between these two. Any idea how that can be done?
You can sample x1 and x2 from 𝑝1(𝑥|𝜇1,Σ1) and 𝑝2(𝑥|𝜇2,Σ2) respectively, then compute KL divergence using torch.nn.functional.kl_div(x1, x2).
As @Nikronic mentioned the kl_div requires a to be log-probabilities and the target b to be probability. correct me if I’m wrong, but I think sampling from the two distributions is not going to give log-probabilities and probabilities
any suggestion? I don’t think that what you said can be used for sampling layer
@Rojin
Actually, I have never used distribution class but based on docs, in this situation you just need to pass them directly to kl_div from distribution if the aforementioned distributions are obtained from Distribution subclasses or you have registered your own.
Please refer to this links for more information:
- https://pytorch.org/docs/stable/distributions.html#torch.distributions.kl.kl_divergence
- https://pytorch.org/docs/stable/distributions.html#torch.distributions.distribution.Distribution
well, you know, my name is Nik, somehow abbreviated of Nikan (a Persian name) 
Bests
Hey Nikan!
 Yeah I realized that from your last name!
Yeah I realized that from your last name!
Sorry if I’m restating my question ( I modified it a couple of times at the top lol) and probably repeating your answer, I’m kinda new to the KL implementation in pytorch, so I really appreciate your help.
My question is :
I have two VAEs, each with a mu and standard deviation layers. Now I would like to calculate the KL between these two VAEs using either the mu and sds, or the sampling layers. This sounds to me like a multivariate gaussian KL divergence problem, so I looked at the formula and I noticed that I actually need the covariance matrix of q (if we assume that KL(p||q) ). And as far as I know there is no way to calculate the covariance in this case, am I correct?
@zhl515 suggested to directly calculate the KL between the two sampling layers as you said before using the bellow function, but as you said this requires probabilities which I do not have, so I think the following function cannot be used on the sampling layers, is that right? :
torch.nn.functional.kl_div(p, q).
And based on your last comment, you are suggesting to register the distribution, and then use
torch.distributions.kl.kl_divergence(p, q)
The only problem is that in order to register the distribution I need to have the covariance matrix, and I can’t obtain that because I only have mu and std. It makes me think whether that is a possible thing to do at all in neural network. In the VAE paper they don’t have that problem because the are assuming that q has mean 0 and covariance I
Thank you!
You said you can’t obtain covariance matrix. In VAE paper, the author assume the true (but intractable) posterior takes on a approximate Gaussian form with an approximately diagonal covariance. So just place the std on diagonal of convariance matrix, and other elements of matrix are zeros.
Now you can compute KL-divargence of two multivariate Gaussians directly from the below formula:
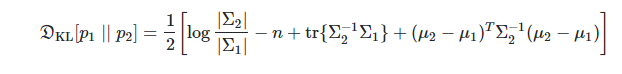
Thank you that solves the problem 
Hey @Rojin
Could you share how you computed the KL-divergence? I am also trying to achieve the same
Hi, @zhl515.
Thanks for your solution and I’ve coded as you post, like this:
def kl_divergence(mu1, mu2, sigma_1, sigma_2):
sigma_diag_1 = np.eye(sigma_1.shape[0]) * sigma_1
sigma_diag_2 = np.eye(sigma_2.shape[0]) * sigma_2
sigma_diag_2_inv = np.linalg.inv(sigma_diag_2)
kl = 0.5 * (np.log(np.linalg.det(sigma_diag_2) / np.linalg.det(sigma_diag_2))
- mu1.shape[0] + np.trace(np.matmul(sigma_diag_2_inv, sigma_diag_1))
+ np.matmul(np.matmul(np.transpose(mu2 - mu1), sigma_diag_2_inv), (mu2 - mu1))
)
return kl
But I have another problem that I cannot call it in a “vectorized” way, or say, I don’t know how to let my code support batch operation in Pytorch. For instance, I have a tensor mu_1 with shape [batch_size,n] and another tensor mu_2 with shape [batch_size,n]. The variance tensors are same with mu. The n here is the dimensions of Gaussian distribution. How can I calculate kl loss for each instance without using for-loop ?
Any ideas? thx
Check this thread.
torch.diag_embed() should do the trick.
hi i don’t know much about the meaning of mu1 mu2 sigma_1 and sigma_2
I am not sure I understand fully your question, but IMO if you have mu1, mu2, sigma_1, sigma_2, why not create two instances of MultivariateNormal using diagonal covariance matrices?
# mu1 [batch_size, n]
# assuming that sigma1 is [batch_size, n] also
cov1 = torch.stack([torch.diag(sigma) for sigma in torch.exp(sigma_1)])
mvn1 = MultivariateNormal(mu1, cov1)
...
# similarly for mvn2
MultivariateNormal will interpret the batch_size as the batch dimension automatically thus mvn1 would have:
batch_shape = batch_size
event_shape = n
sample_shape = ()
when you sample it will take into consideration the batch_shape.
You can compute kl (mvn1, mvn2) using the Pytorch’s implementation.
I hope this can help.
This is really helpful!
Thanks a lot.
Is this formula applicable to any arbitrary distribution? e.g., non-Gaussian?
The cov-variance matrix must be positive definite.
Any idea how to turn a diagonal matrix with zero non-diagonal elements into a positive definite matrix?
The output comes from an FC+BN+ReLU layer.
then I can turn it into a diagonal by:
cov_var_diag = torch.diag_embed(output)
cov_var_spd = cov_var @ cov_var.mT + 1e-6 # this actually makes symmetric positive definite and didn't work
q = torch.distributions.multivariate_normal.MultivariateNormal(loc=mu, covariance_matrix=cov_var_spd)
Also looked into Cholesky decomposition into lower triangular matrix as scale_tril attribute in MVN function, but that also didn’t end well.