Hi,
I am training a simple attention network with stored extracted ResNet features. Every gigapixel image is divided into approximately 20000 patches of size 256x256, and each patch is associated with a feature vector from custom ResNet50. Now, my shape of data for every image will be [20000, 1024].
The train data loader loads a gigapixel image at a time, making the batch size 1.
Reference: https://github.com/mahmoodlab/CLAM
Model
class Attn_Net_Gated(nn.Module):
def __init__(self, L = 1024, D = 256, dropout = False, n_classes = 1):
super(Attn_Net_Gated, self).__init__()
self.attention_a = [
nn.Linear(L, D),
nn.BatchNorm1d(D),
nn.Tanh()]
self.attention_b = [nn.Linear(L, D),
nn.BatchNorm1d(D),
nn.Sigmoid()]
if dropout:
self.attention_a.append(nn.Dropout(0.25))
self.attention_b.append(nn.Dropout(0.25))
self.attention_a = nn.Sequential(*self.attention_a)
self.attention_b = nn.Sequential(*self.attention_b)
self.attention_c = nn.Linear(D, n_classes) #Linear(L,D); L - input, D - output
def forward(self, x):
a = self.attention_a(x)
b = self.attention_b(x)
A = a.mul(b)
A = self.attention_c(A)
return A, x
class MB(nn.Module):
def __init__(self, gate = True, size_arg = "small", dropout = False, k_sample=8, n_classes=2,
instance_loss_fn=nn.CrossEntropyLoss(), subtyping=True):
nn.Module.__init__(self)
self.size_dict = {"small": [1024, 512, 256], "big": [1024, 512, 384]} #choosing the model size
size = self.size_dict[size_arg]
fc =[]
if gate:
attention_net = Attn_Net_Gated(L = size[0], D = size[2], dropout = dropout, n_classes = n_classes)
fc.append(attention_net)
self.attention_net = nn.Sequential(*fc)
self.n_classes = n_classes
self.subtyping = subtyping
initialize_weights(self)
def relocate(self):
device=torch.device("cuda" if torch.cuda.is_available() else "cpu")
def forward(self, h, label=None, instance_eval=False, return_features=False, attention_only=False):
device = h.device
A, h = self.attention_net(h)
A = torch.transpose(A, 1, 0)
if attention_only:
return A, h
Utils
def get_split_loader(split_dataset, training = False, testing = False, weighted = False):
"""
return either the validation loader or training loader
"""
kwargs = {'num_workers': 4} if device.type == "cuda" else {}
if not testing:
if training:
if weighted:
weights = make_weights_for_balanced_classes_split(split_dataset)
loader = DataLoader(split_dataset, batch_size=1, sampler = WeightedRandomSampler(weights, len(weights)), collate_fn = collate_MIL_tr, **kwargs)
else:
loader = DataLoader(split_dataset, batch_size=1, sampler = RandomSampler(split_dataset), collate_fn = collate_MIL_tr, **kwargs)
else:
loader = DataLoader(split_dataset, batch_size=1, sampler = SequentialSampler(split_dataset), collate_fn = collate_MIL_tr, **kwargs)
else:
ids = np.random.choice(np.arange(len(split_dataset), int(len(split_dataset)*0.1)), replace = False)
loader = DataLoader(split_dataset, batch_size=1, sampler = SubsetSequentialSampler(ids), collate_fn = collate_MIL_tr, **kwargs )
return loader
def get_optim(model, args):
if args.opt == "adam":
optimizer = optim.Adam(filter(lambda p: p.requires_grad, model.parameters()), lr=args.lr, weight_decay=args.reg)
elif args.opt == 'sgd':
optimizer = optim.SGD(filter(lambda p: p.requires_grad, model.parameters()), lr=args.lr, momentum=0.9, weight_decay=args.reg)
else:
raise NotImplementedError
return optimizer
def initialize_weights(module):
for m in module.modules():
if isinstance(m, nn.Linear):
nn.init.xavier_normal_(m.weight)
m.bias.data.zero_()
elif isinstance(m, nn.BatchNorm1d):
nn.init.constant_(m.weight, 1)
nn.init.constant_(m.bias, 0)
Training
def train(datasets, cur, args):
train_split, val_split, test_split = datasets
save_splits(datasets, ['train', 'val', 'test'], os.path.join(args.results_dir, 'splits_{}.csv'.format(cur)))
model = MB(**model_dict, instance_loss_fn=instance_loss_fn)
model.relocate()
optimizer = get_optim(model, args)
train_loader = get_split_loader(train_split, training=True, testing = args.testing, weighted = args.weighted_sample)
val_loader = get_split_loader(val_split, testing = args.testing)
test_loader = get_split_loader(test_split, testing = args.testing)
if args.early_stopping:
print('yes')
early_stopping = EarlyStopping(patience = 20, stop_epoch=50, verbose = True)
else:
early_stopping = None
for epoch in range(args.max_epochs):
if args.model_type in [''mmb'] and not args.no_inst_cluster:
epoch_loss = train_loop(epoch, model, train_loader, optimizer, args.n_classes, args.bag_weight, writer, loss_fn)
stop, val_loss = validate(cur, epoch, model, val_loader, args.n_classes, early_stopping, writer, loss_fn, args.results_dir)
if stop:
break
if args.early_stopping:
model.load_state_dict(torch.load(os.path.join(args.results_dir, "s_{}_checkpoint.pt".format(cur))))
else:
torch.save(model.state_dict(), os.path.join(args.results_dir, "s_{}_checkpoint.pt".format(cur)))
return epoch_loss, val_loss
def train_loop(epoch, model, loader, optimizer, n_classes, bag_weight, writer = None, loss_fn = None):
device=torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.train()
epoch_loss = 0.
for batch_idx, (data, label, coordinates, slide_id) in enumerate(loader):
data, label = data.to(device), label.to(device)
pred_val, h_feat = model(data, label = label, attention_only = True)
target_val = #this is an array that I obtain from some interpolation and has the same shape of pred_val
l2_loss = coeff * torch.nn.functional.mse_loss(pred_val.unsqueeze(0), target_val.unsqueeze(0))
epoch_loss += l2_loss.item()
optimizer.zero_grad()
l2_loss.backward()
optimizer.step()
epoch_loss = epoch_loss / len(loader)
print('Epoch: {}, train_loss: {:.4f} '.format(epoch, epoch_loss))
return epoch_loss
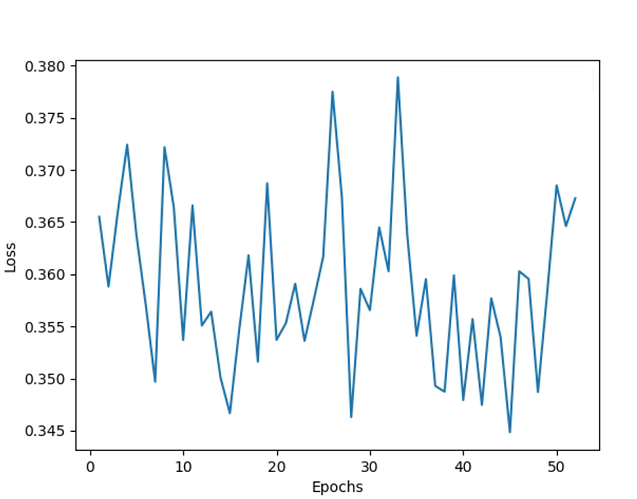
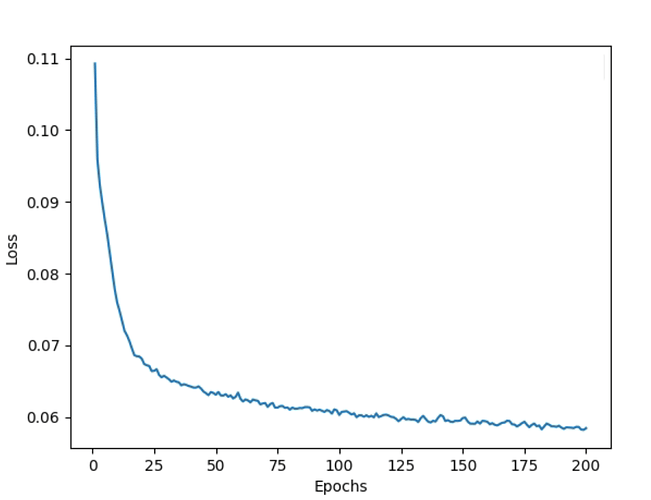
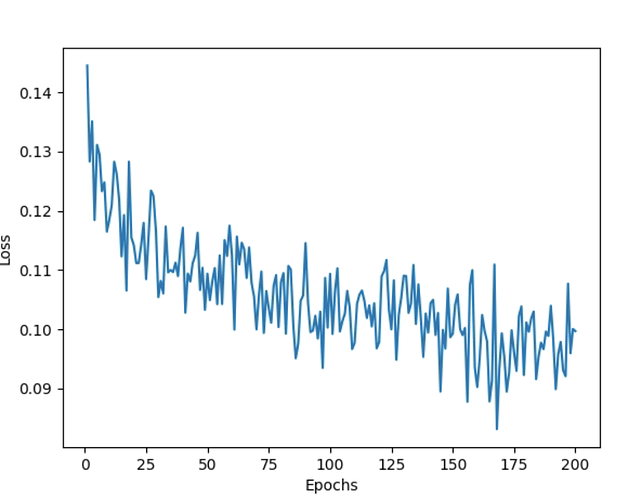
The train loss oscillates and gets stuck within a fixed range of values as follows and does not minimize:
NOTE: I have tried the above experiments for Learning rates ranging from 1e-2 to 1e-6; Weight decay from 1e-3 to 1e-6; for optimizers both Adam and SGD; epochs from 50 to 200 (with and without early stopping). The loss graph for all the experiments conducted so far is similar to the above snapshot.
Any help is appreciated.