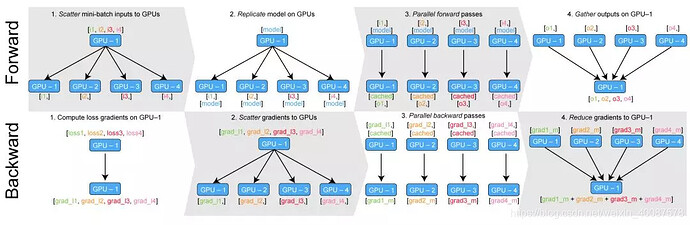
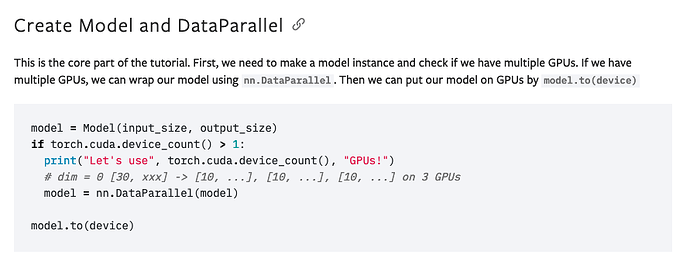
Another weird problem in nn.DataParallel
in my main.py I put the model to device
encoder = Encoder(protein_dim, hid_dim, n_layers, kernel_size, dropout)
decoder = Decoder(atom_dim, hid_dim, n_layers, n_heads, pf_dim, DecoderLayer, SelfAttention,
PositionwiseFeedforward, dropout)
model = Predictor(encoder, decoder)
# model.load_state_dict(torch.load("output/model/lr=0.001,dropout=0.1,lr_decay=0.5"))
model = nn.DataParallel(model)
model.to(device)
trainer = Trainer(model, lr, weight_decay, scaler)
tester = Tester(model)
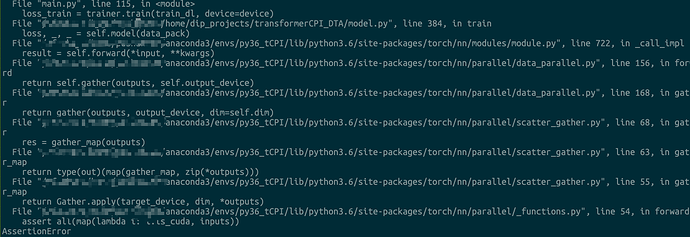
loss_train = trainer.train(train_dl, device=device) # This line throw errors
But I got the following error
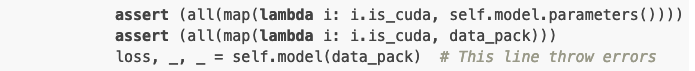
assert all(map(lambda i: i.is_cuda, inputs))
AssertionError
I have test all model.parameters() and inputs in train():
def train(self, dataloader, device):
self.model.train()
if self.scaler is None:
for i, data_pack in enumerate(dataloader):
data_pack = to_cuda(data_pack, device=device)
assert (all(map(lambda i: i.is_cuda, self.model.parameters())))
assert (all(map(lambda i: i.is_cuda, data_pack)))
loss, _, _ = self.model(data_pack) # This line throw errors
self.optimizer.zero_grad()
loss.sum().backward()
self.optimizer.step()
The results are all True. But I still get this error in the third line loss, _, _ = self.model(data_pack)
What happened?
This is my forward function:
def forward(self, data):
compound, adj, protein, correct_interaction, atom_num, protein_num = data
# compound = [batch,atom_num, atom_dim]
# adj = [batch,atom_num, atom_num]
# protein = [batch,protein len, 100]
compound_max_len = compound.shape[1]
protein_max_len = protein.shape[1]
compound_mask, protein_mask = self.make_masks(atom_num, protein_num, compound_max_len, protein_max_len)
compound = self.gcn(compound, adj)
# compound = torch.unsqueeze(compound, dim=0)
# compound = [batch size=1 ,atom_num, atom_dim]
# protein = torch.unsqueeze(protein, dim=0)
# protein =[ batch size=1,protein len, protein_dim]
enc_src = self.encoder(protein)
# enc_src = [batch size, protein len, hid dim]
predicted_interaction = self.decoder(compound, enc_src, compound_mask, protein_mask)
# out = [batch size, 2]
# out = torch.squeeze(out, dim=0)
loss = self.Loss(predicted_interaction, correct_interaction.view(-1, 1))
return torch.unsqueeze(loss, 0), predicted_interaction.cpu().detach().view(-1, 1), correct_interaction.cpu().detach().view(-1, 1)
Thank you very much !!!