@ptrblck what else should I check other than grad_fn ?
Once all .grad_fn = None issues are solved, you could verify that all trainable parameters get valid gradients after the first backward() call by checking their .grad attribute:
loss.backward()
for name, param in model.named_parameters():
print(name, param.grad)
If you are seeing gradients for the expected parameters and the model is still not training, try to overfit a tiny dataset (e.g. just 10 samples) by playing around with some hyperparameters.
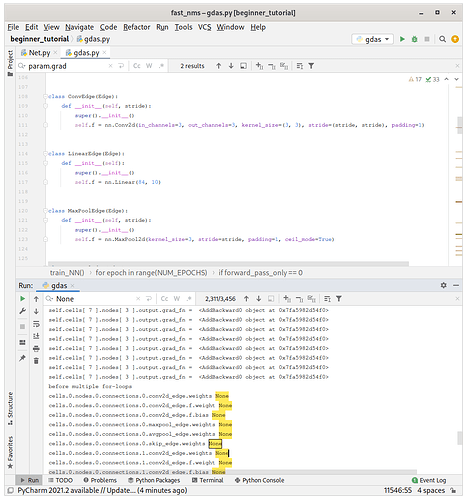
Strange, this still prints out None …
This would indicate, that the detaching issue is not solved yet or these parameters were never used in the computation graph.
If they had been instantiated with such hierarchy : cells.0.nodes.0.connections.0.conv2d_edge.weights , why are they still not used in the computation graph ?
Strange…
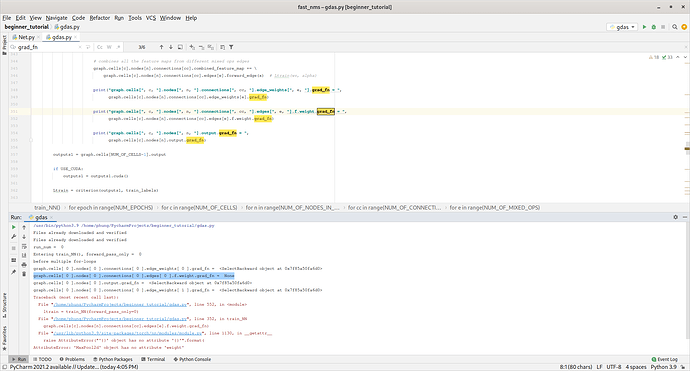
@ptrblck wait, I just found that there are still grad_fn = None issues for conv2d_edge which is also labelled as edges[0].f.weight
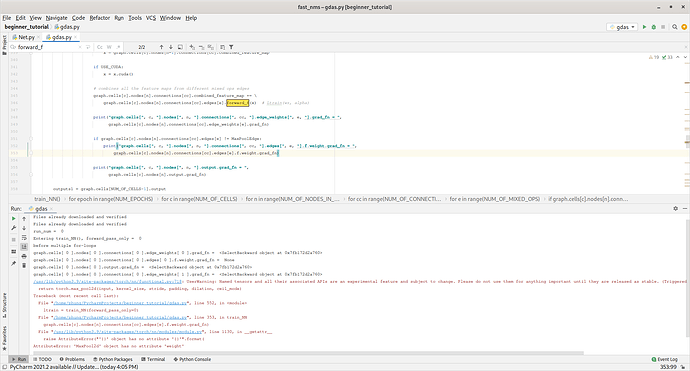
You’re just calling forward_edge, but you’re actual conv layers get called in forward_f.
I found a strange line of code which might had led to f.weight.grad = None
forward_f(x) does not really call the self.f inside class ConvEdge(Edge)
# for NN functions internal weights training
def forward_f(self, x):
self.__unfreeze_f()
self.__freeze_w()
# inheritance in python classes and SOLID principles
# https://en.wikipedia.org/wiki/SOLID
# https://blog.cleancoder.com/uncle-bob/2020/10/18/Solid-Relevance.html
return self.f(x)
# self-defined initial NAS architecture, for supernet architecture edge weight training
def forward_edge(self, x):
self.__freeze_f()
self.__unfreeze_w()
return x * self.weights
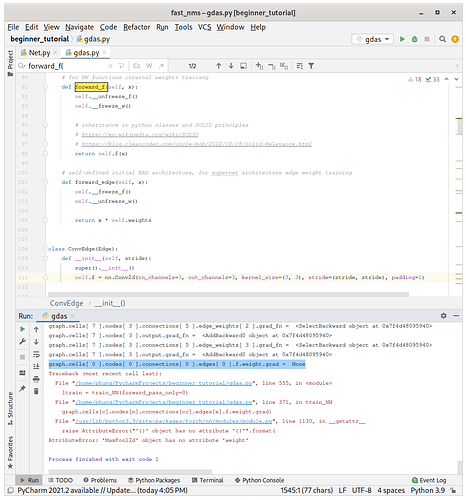
class ConvEdge(Edge):
def __init__(self, stride):
super().__init__()
self.f = nn.Conv2d(in_channels=3, out_channels=3, kernel_size=(3, 3), stride=(stride, stride), padding=1)
One problem is that you do
outputs1 = graph.cells[NUM_OF_CELLS-1].output
Ltrain = criterion(outputs1, train_labels)
but as far as I can see, outputs1 isn’t used in the computation graph we’re interested in.
outputs1 is the output of the computation graph, of course it is not used inside the computation graph.
What is wrong with outputs1 then ?
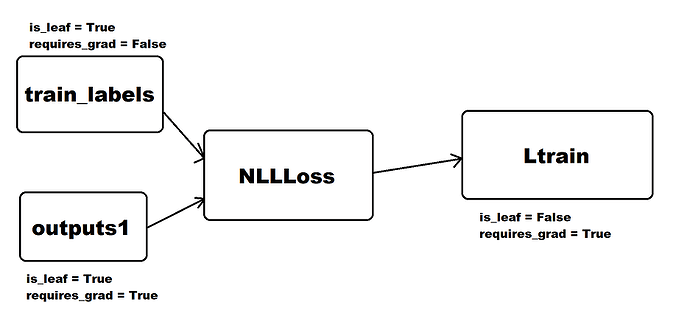
I’ve tried to visualize the computation graph what I think you’re backwarding through:
I hope this visualization helps, if I haven’t overlooked anything.
it seems the picture tells it all, but do I need requires_grad = True for train_labels ?
By the way, how does this picture tell why f.weight.grad = None occur ?
That is because, as you can see in the picture, the weight is not in the computation graph you’re backwarding.
Furthermore, by checking the attributes of outputs1, you can also see that it still is the user created tensor.
outputs1 comes from the computation results of self.cells[c].nodes[n].output inside class Graph.
and I noticed that these computations make use of edge_weights and does not make use of f.weight
outputs1 comes from graph.cells[NUM_OF_CELLS-1].output, doesn’t it? And it is the same tensor as the one you’ve initialized.
I solved all the concern you raised regarding outputs1.
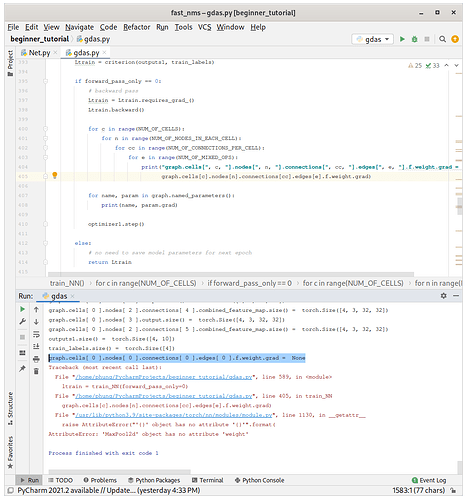
However, the code still give graph.cells[ 0 ].nodes[ 0 ].connections[ 0 ].edges[ 0 ].f.weight.grad = None
Yeah, I’ve checked the computation graph, which should be fine for some cells.
I think you therefore should recheck the GDAS specific logic happening there.
Furthermore, you’re declaring c multiple times, is this intentional?
Which exact line of coding did I declare c multiple times ?
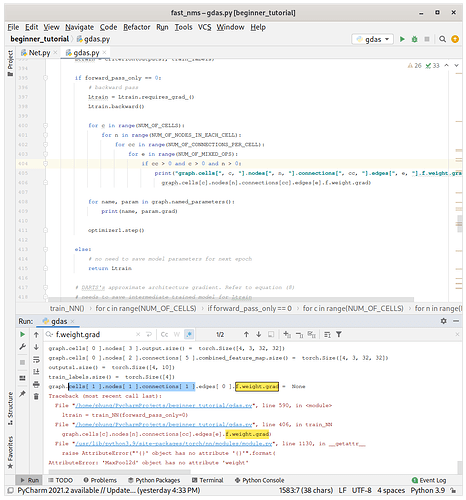
it is not just for 0 combination, it happens to other combinations as well.
For instance at line 336 and line 366 in your current code.