Since I got a couple of questions in this previous thread, which aims to order sequence data into batches where all input sequences in a batch have the same length. This avoids the need of padding and optional packing.
The original solution work only for sequence classification, sequence tagging, autoencoder models since the ordering only considered the input sequences. For Seq2Seq models such as machine translation, where the output sequences also differ in lengths, it didn’t work.
I’ve now made the small changes to support this as well – here is the code for the Sampler and my own Vectorizer I use, as well as a Jupyter Notebook showing the usage. Now, each batch contains sequence pairs where all input sequences are the same length and all target sequences have the same length. For example a batch might contain 32 pairs with all input sequences having length 12 and all target sequences having length 15.
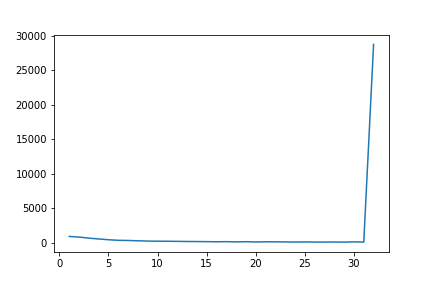
To see how the combination of input and target lengths are distributed, I’ve made a test with a real-world dataset for machine translation. I’ve set the batch size to 32. The figure below shows the distribution of the sizes of all batches. As one can see, the vast majority of batches is indeed full (i.e., 32 sequence pairs). This shouldn’t really be surprising since:
- Batch sizes (e.g., 32 or 64) are is essentially nothing given large datasets of millions of sequences pairs or more. Thus, the chance that enough pairs share the same input and target lengths is high.
- The combination of input and target lengths is typically not independent. For example, an input of length 5 generally does not have a target length of 20 but in a similar range.
In short, I think it’s a convenient solution for working with batches in case of Seq2Seq models, again, with no need for padding and packing – and way faster that training with a batch size of 1. Maybe it’s useful for some of you.
